The connection between Darwin’s finches and bacterial flagellar motors
DOI: 10.1063/pt.nkma.bqid
Watching bacteria dance in a droplet of water under a microscope is an awe-inspiring experience that evokes emotions of wonder, like those one might feel while observing stars in the sky. Bacteria’s motility is the result of billions of years of evolution, and in many species, it is driven by a glorious macromolecular structure known as the flagellum. That biological masterpiece consists of multiple distinct parts, as seen in figure
Figure 1.

A swimming bacterium uses a flagellum to power its motion. A slice through a cryo-electron tomogram of a Shewanella oneidensis cell illustrates the filament, motor, and hook that make up the appendage.
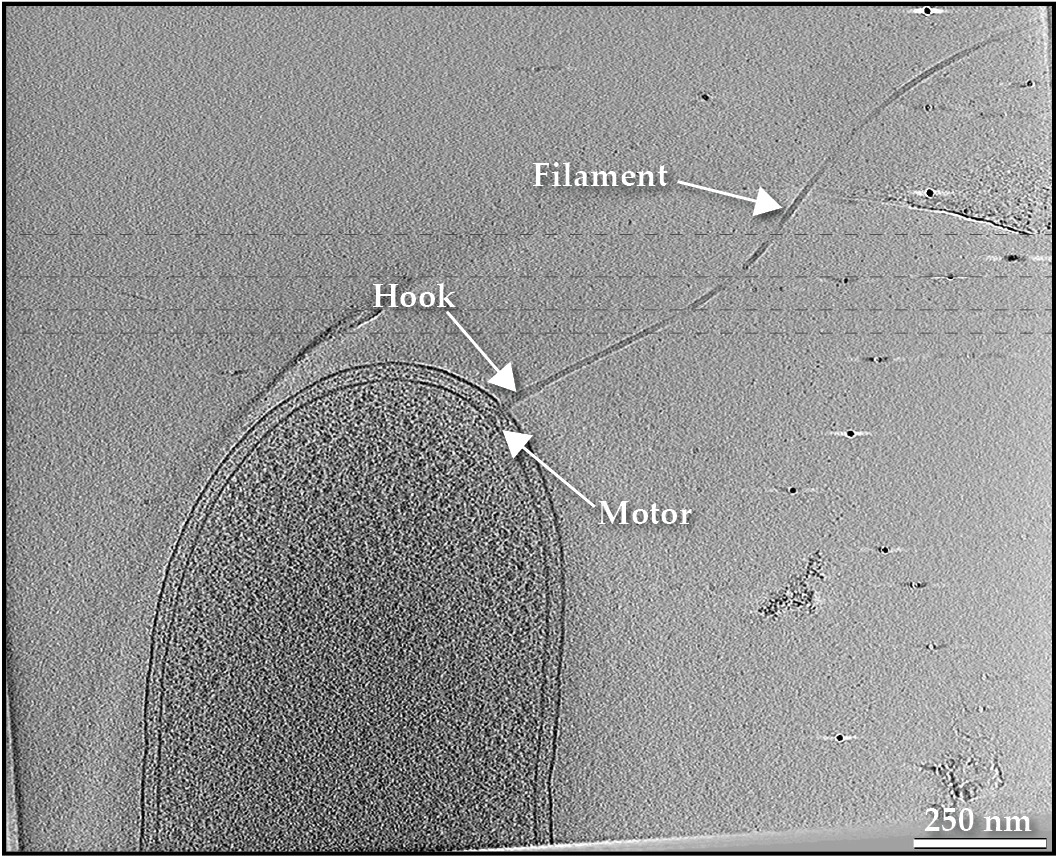
The structure, function, and assembly of the motor have long captured the imagination of both academic scholars and science popularizers (see the article by Howard Berg, Physics Today, January 2000, page 24
In 1835 Charles Darwin arrived at the Galápagos Islands during the course of his famous voyage aboard the Beagle. There he studied many biological species, including what became known as Darwin’s finches, the birds that inhabit different areas of the islands. Those birds share a common ancestor—the dull-colored grassquit—that lives on mainland South America, but they have evolved a remarkable variety of beak types to suit their characteristic, area-specific food chains. For example, birds with blunt beaks usually feed on seeds, while those with long and sharp beaks tend to hunt for and grab insects. They represent iconic instances of evolution and adaptation to the environment. In a certain way, the same is true for bacterial flagellar motors.
All bacterial flagellar motors discovered to date have similar shared core structures. Several bacteria species, however, have developed embellishments that optimize the motor to suit a specific environmental niche. For instance, species that inhabit more viscous environments have enhanced motors with extra components to generate higher torque. The diversity of flagellar motors is a prime example of the ongoing evolution of macromolecular biological machines, comparable to the diverse beaks of Darwin’s finches. The flagellar motor characteristic of a given species can, in fact, be viewed as a “molecular fossil,” and every time the architecture of a different motor type is revealed, a new tile clicks into the jigsaw puzzle of how those amazing nanomachines have evolved over time.
For the sake of simplicity, the scope of this article is limited to bacteria with a cell envelope containing inner and outer membranes. All slices of cryo-electron tomograms or subtomogram averages of flagellar motors depicted here represent 2D cross sections through 3D reconstructions of the protein complexes. Therefore, a cross section through a ring appears as two dots (for example, the bushings in figure
Figure 2.

The Escherichia coli flagellar motor is made of many components that play similar roles to the standard parts of a mechanical motor. A central slice through a subtomogram average of the flagellar motor (left) is overlaid with labeled component parts (right).
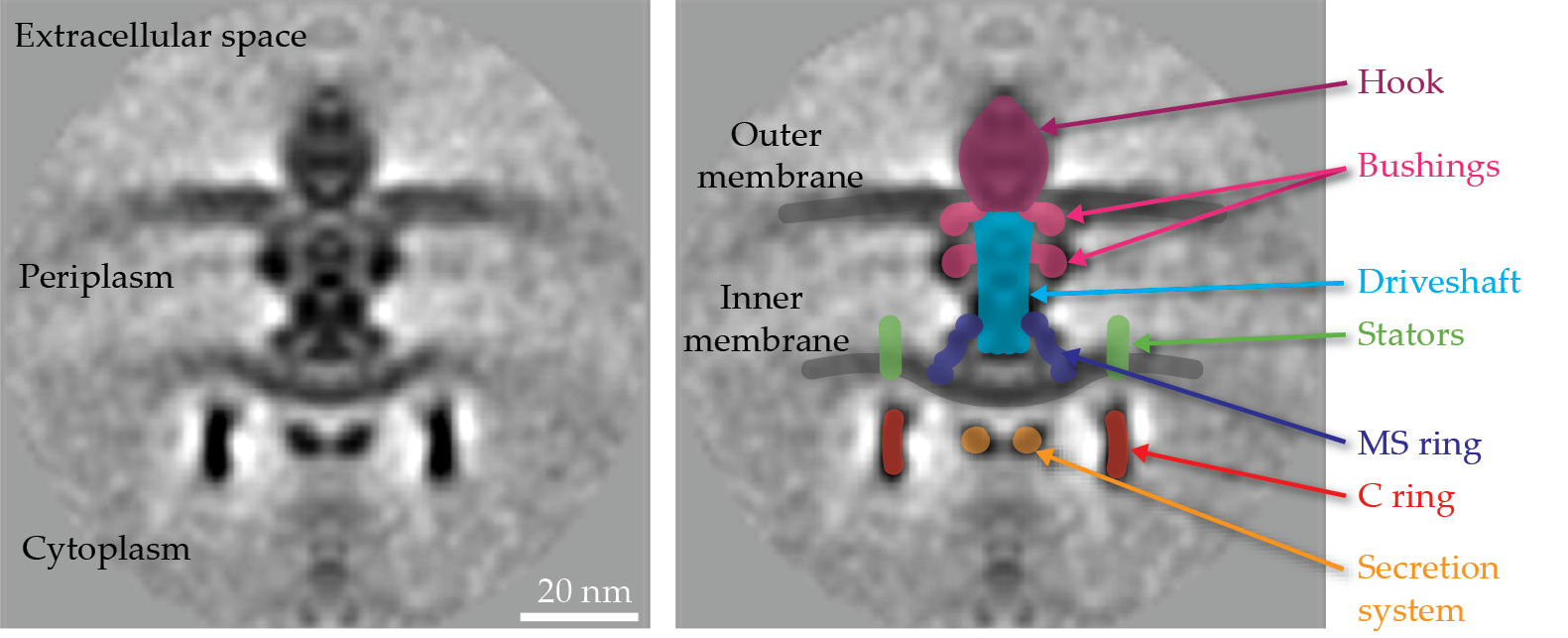
Anatomy of a flagellar motor
In his book The God Delusion, Richard Dawkins wrote of the flagellar motor, “It derives the only known example, outside human technology, of a freely rotating axle.” The components of a flagellar motor, like the Escherichia coli motor seen in figure
The rotor comprises a series of rings, including the cytoplasmic ring, known as the C ring, and the inner-membrane embedded ring, or the MS ring. A dedicated secretion system delivers proteins—used to build the driveshaft, hook, and filament—across the inner membrane to the periplasmic space (the space between the inner and outer membranes, shown in figure
Stators, which are ion channels embedded in the inner membrane and surround the rotor, generate torque by using ion motive force produced by electrical and chemical potential differences across the cell membrane. Recent work has revealed the mechanism of stator–rotor interaction that generates the torque and drives both cell motility and a rotational switching mechanism.
1–3
Stators appear to operate like smaller rotors themselves, spinning as they pump ions across the inner membrane and interacting with the upper part of the C ring to drive its rotation and thereby generate torque, as shown in figure
Figure 3.

Rotational switching of the flagellar motor is accomplished by changes to the C ring’s shape and position, which are controlled by a cytoplasmic protein (CheY-P). Stators always spin clockwise, so when they engage the outer surface of the C ring (left), the motor spins counterclockwise. When stators engage the inner surface of the C ring (right), the motor spins clockwise.
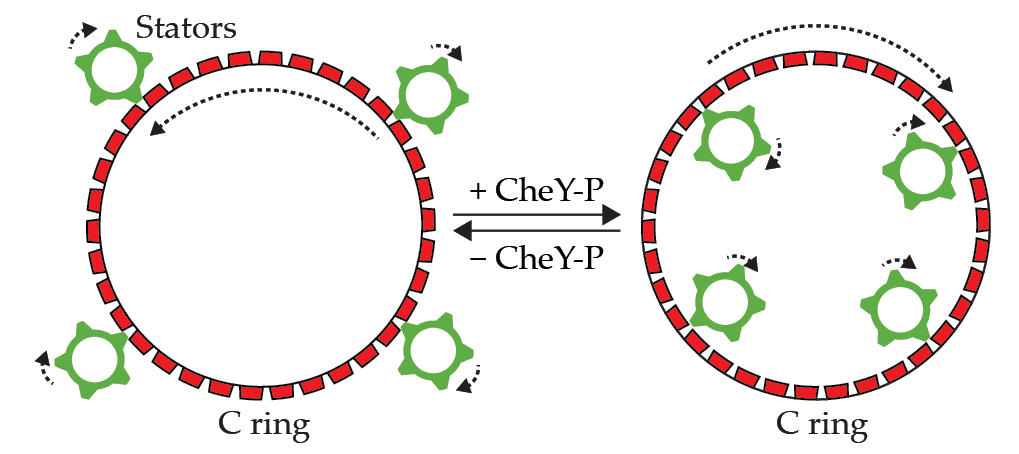
While stators always spin clockwise, the motor can alternate between spinning clockwise (CW) and counterclockwise (CCW) by using a clever trick. When the outer surface of the C ring is touching the CW-spinning stators, the C ring spins in the direction counter to that of the stators, and the motor rotates CCW, as shown in the left panel of figure
Macromolecular structural diversity
The structure of the motor outlined in figure
Just like every bird type of Darwin’s finches evolved a beak that suits the food sources available in its milieu, every bacterial species evolved its characteristic macromolecular motor structure to suit the environmental niche it inhabited.
6
,
7
For instance, the motor of the pathogen Campylobacter jejuni, shown in the
Seeing inside bacteria with cryo-ET
In cryo-electron tomography, or cryo-ET, cells are plunge-frozen on an electron microscope grid using an efficient coolant, which prevents the formation of ice crystals and instead promotes the development of vitreous (noncrystalline) ice. Subsequently, a transmission electron microscope takes a series of images at different tilt angles as the sample is rotated. Typically, images are taken every 1° to 3°. The 2D images are then computationally combined to build a 3D reconstruction of the sample at a resolution of a few nanometers, which allows for the visualization of macromolecular complexes inside the individual, intact hydrated-frozen cells.

The cryo-ET data collection method used to produce images of bacteria is illustrated in the upper panel. A slice through a cryo-electron tomogram of a Campylobacter jejuni cell depicts the different parts of its flagellum in the lower left panel. A central slice through the subtomogram average of a C. jejuni flagellar motor,
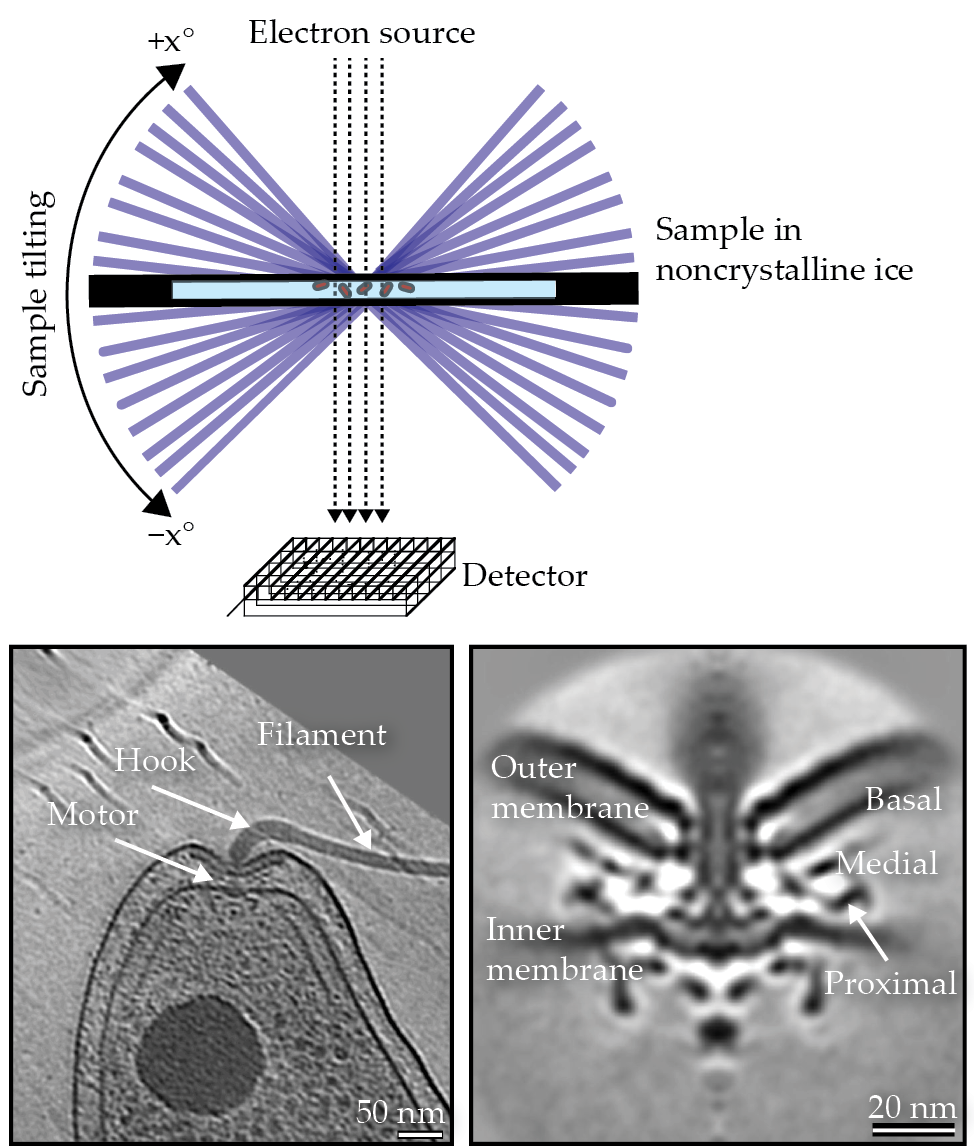
The overall resolution can be improved by performing a subtomogram averaging procedure, wherein fragments of the tomogram containing a complex of interest—for example, a bacterial flagellar motor—are computationally identified, cropped, aligned, and averaged together.
The power of cryo-ET lies in its ability to produce images of macromolecular complexes in their native cellular milieu, without the need to purify or perturb them, and at a macromolecular resolution. In other words, its strength lies in avoiding the specific concerns that Johann Wolfgang von Goethe expressed in Faust:
When scholars study a thing, they strive
To kill it first, if its alive;
Then they have the parts and they’ve lost the whole,
For the link that’s missing was the living soul.
A crucial point about evolution is that it is not always progressive. Although it sometimes leads to increased complexity, traits can also be lost during the evolutionary process if that loss proves beneficial for the species. 8 Therefore, just because E. coli has the bare conserved-core motor does not necessarily mean that its motor is “older” or “ancestral” to others. In fact, recent work has shown that the E. coli motor is not native to that particular species but, rather, has been acquired from other bacteria through gene-transfer mechanisms. 9 That turns out to be advantageous because the generic motor easily adjusts to suit the various environments that E. coli commonly encounters in its niche, such as in the gut.
Structural diversity is not limited to the motor alone. For example, spirochaetes evolved endoflagella—or periplasmic flagella—where the hook and the filament remain in the periplasm. That configuration, with the filament wrapping around the cell beneath the outer membrane, results in the bacteria moving by rolling or undulating in highly viscous, gel-like environments. Other species, including many pathogens, have what is called “sheathed flagella,” with the outer membrane extending to surround the hook and the filament. The way by which the sheath forms, and its exact function, remains enigmatic, but it could be a way for the bacterium to circumvent the host immune system by sequestering the flagellar filament protein inside the sheath to prevent a reaction to that protein by the host. 10
Building a flagellar motor
The process by which a bacterial cell assembles a flagellar motor has long fascinated scientists. How do individual proteins of all kinds assemble into such an intricate nanomachine? Since the flagellar motor consumes a significant amount of precious cell energy, its assembly is tightly regulated. Its biogenesis is, in a sense, inside out: Components associated with the interior of the cell tend to assemble first, and the process propagates upward through the periplasmic space and on to the outer membrane,
11
as shown in figure
Figure 4.

Assembly stages of a flagellar motor in a Hylemonella gracilis cell. The highlighting in the tomographic slices shows the addition of motor components.
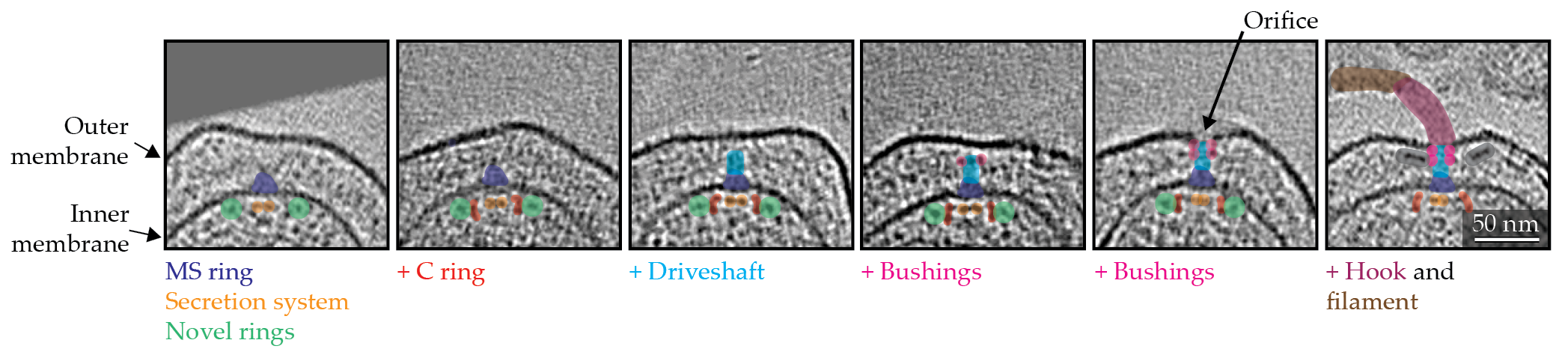
At the beginning of the process, the inner-membrane components assemble, and, subsequently, the driveshaft proteins click into place. On completion of the driveshaft biogenesis, the bushings are built around it. The formation of the bushings reshapes the outer membrane to create an orifice, thereby allowing the assembly of the extracellular hook and filament.
The above process, however, is not merely an accretion of the component proteins. It appears to be dynamic, with various chaperones and enzymes involved. Chaperones are proteins that help with folding and translocating other proteins to their cellular locale. Recent intracellular cryo-ET imaging of the motor biogenesis process has revealed transient cytoplasmic rings (shown as green circles in figure
The biogenesis processes of various embellishments to the conserved-core motor are not yet fully understood. In one interesting case though, an extra periplasmic disk in the bacterial species Wolinella succinogenes was observed assembling in a manner akin to an Archimedean spiral, with a protein polymerizing to build a spiral around the bushings. 13 Future observations may reveal equally novel construction mechanisms among those embellishments.

KTSDESIGN/SHUTTERSTOCK.COM
Disassembly processes
Whereas the process of flagellar assembly has been under scrutiny for some 30 years, the fact that bacteria lose their flagella—and even disable the motor at times—is a recent discovery, independently reported by multiple research groups a few years ago. 14–18 One such form of flagellar loss occurs during starvation. Because the flagellum is an energetically expensive nanomachine, bacteria may eject their flagellar filaments, including hooks, when they encounter an environment with sparse nutrients.
Interestingly, on ejecting the extracellular protrusions, the motor is partially disassembled, and only the bushing rings remain as a relic in the outer membrane, readily observable using cryo-ET. Whereas the bushings appear to be hollow when surrounding the driveshaft before the flagellar loss, they become plugged when the relic rings are left behind. The process of flagellar loss is observed throughout a vast variety of bacterial species, suggesting that it is inherent to flagellar motors. Thus, in addition to acting as bushings during the course of normal operation, the plugged bushing rings may play a significant role in keeping the membrane intact by sealing up the hole following a flagellar loss event under starvation conditions. 14
The process of ejecting the extracellular protrusions and forming the bushing ring plug, however, is not the only way by which bacteria can lose their flagella and disable the motor. For instance, it has been demonstrated that under severe stress, such as cell disintegration, the motor can lose its C ring, 15 which suggests a weak interaction between the C ring and the rest of the motor.
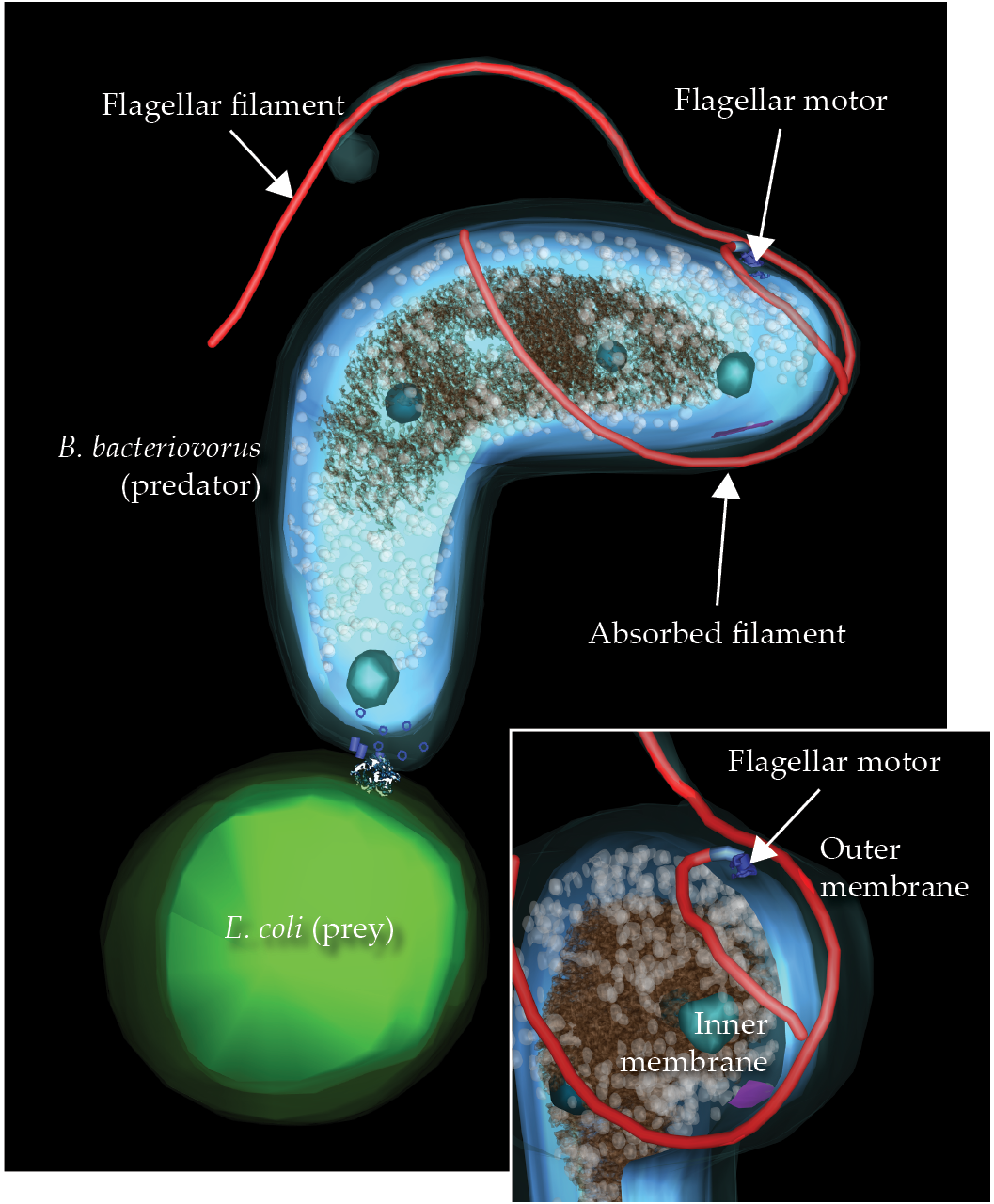
One spectacular flagellar-loss process occurs in the microbial predator Bdellovibrio bacteriovorus. To consume other bacteria, it enters the periplasmic space between the prey’s inner and outer membranes. As shown in figure
Figure 5.

A predator bacteria cell, Bdellovibrio bacteriovorus, is in the process of capturing an Escherichia coli minicell. When catching a prey bacterium, B. bacteriovorus retracts its extracellular hook and filament into its own periplasmic space, shifting the motor to the side to make room for the retraction process.
Evolving molecular nanomachines
Humans have always been fascinated by both the smallest- and largest-scale worlds. In Jonathan Swift’s classic Gulliver’s Travels, Gulliver encounters both worlds in his voyages to Lilliput and Brobdingnag. And now techniques such as cryo-ET have transformed the invisible world of microbes into a Lilliputian environment for researchers, much the same way that observatories and telescopes have transformed the phenomena in outer space into beautiful worlds for astronomers. The propeller-like motion driven by the flagellar motor is but one form of motion used by microbes because different species are known to propel themselves in different ways by using various molecular nanomachines. For example, some cells use another set of nanomachines known as pili, hair-like appendages that extend and retract to pull the cell over surfaces. Other bacteria glide by moving thin protein filaments that protrude from the cell on special periplasmic tracks using motors embedded in the cell membranes.
Studying the diverse range of nanomachines and macromolecular structures that enable bacterial motility in greater detail and how they assemble, disassemble, and function sheds light on their evolution. In fact, a similar approach has been used to reconstruct life history from the fossil record. It is said that on the sight of a single bone relic, or even a single piece of a bone, Georges Cuvier, the founding father of paleontology, could “recognize and reconstruct the portion of the whole from which it would have been taken.” During the growth of a human embryo, many developmental stages reflect vestiges of the evolutionary history of Homo sapiens, such as the development of embryonic tail-like structures and pharyngeal arches. Similarly, the assembly intermediates and disassembly relics of macromolecular nanomachines keep traces of their past and help scientists reconstruct their history.
I am grateful to Dmitry Shorokhov for useful comments on the manuscript.
References
1. Y. Chang et al., Nat. Struct. Mol. Biol. 27, 1041 (2020). https://doi.org/10.1038/s41594-020-0497-2
2. J. C. Deme et al., Nat. Microbiol. 5, 1553 (2020); https://doi.org/10.1038/s41564-020-0788-8
M. Santiveri et al., Cell 183, 244 (2020). https://doi.org/10.1016/j.cell.2020.08.0163. H. Hu et al., Trends Biochem. Sci. 47, 160 (2022). https://doi.org/10.1016/j.tibs.2021.06.005
4. F. M. Rossmann et al., Mol. Microbiol. 114, 443 (2020). https://doi.org/10.1111/mmi.14525
5. S. Chen et al., EMBO J. 30, 2972 (2011). https://doi.org/10.1038/emboj.2011.186
6. M. Beeby et al., Proc. Natl. Acad. Sci. USA 113, E1917 (2016). https://doi.org/10.1073/pnas.1518952113
7. M. Kaplan et al., eLife 8, e43487 (2019). https://doi.org/10.7554/eLife.43487
8. N. A. Johnson, D. C. Lahti, D. T. Blumstein, Evol. Educ. Outreach 5, 128 (2012). https://doi.org/10.1007/s12052-011-0381-y
9. J. L. Ferreira et al., Front. Microbiol. 12, 643180 (2021). https://doi.org/10.3389/fmicb.2021.643180
10. J. Chu, J. Liu, T. R. Hoover, Biomolecules 10, 363 (2020). https://doi.org/10.3390/biom10030363
11. R. M. Macnab, Annu. Rev. Microbiol. 57, 77 (2003). https://doi.org/10.1146/annurev.micro.57.030502.090832
12. M. Kaplan et al., EMBO J. 41, e109523 (2022). https://doi.org/10.15252/embj.2021109523
13. H. Engelhardt, S. C. Schuster, E. Baeuerlein, Science 262, 1046 (1993). https://doi.org/10.1126/science.8235620
14. M. Kaplan et al., Proc. Natl. Acad. Sci. USA 117, 8941 (2020); https://doi.org/10.1073/pnas.1916935117
M. Kaplan et al., J. Mol. Biol. 433, 167004 (2021). https://doi.org/10.1016/j.jmb.2021.16700415. M. Kaplan et al., mBio 12, e00298-21 (2021). https://doi.org/10.1128/mBio.00298-21
16. M. Kaplan et al., Nat. Microbiol. 8, 1267 (2023). https://doi.org/10.1038/s41564-023-01401-2
17. J. L. Ferreira et al., PLOS Biol. 17, e3000165 (2019); https://doi.org/10.1371/journal.pbio.3000165
M. Kaplan et al., EMBO J. 38, e100957 (2019). https://doi.org/10.15252/embj.201810095718. S. Zhu et al., J. Bacteriol. 201, e00117 (2019); https://doi.org/10.1128/JB.00117-19
X.-Y. Zhuang et al., Mol. Microbiol. 114, 279 (2020). https://doi.org/10.1111/mmi.14511
More about the authors
Mohammed Kaplan is an assistant professor of microbiology at the University of Chicago.





