A folding protein gets caught in the act
DOI: 10.1063/PT.3.4314
To transform from linear chains to three-dimensional structures, in vivo proteins somehow navigate tortuous free-energy landscapes. Their final configurations must be just right for them to function properly; protein misfolding is thought to underlie some allergies as well as neurodegenerative diseases such as Parkinson’s and Alzheimer’s.
X-ray crystallography and NMR are well-established methods for accessing the detailed structure of a protein’s final folded configuration. Gathering dynamical information about the folding process itself requires real-time techniques such as fluorescence, circular dichroism, and hydrogen exchange; acquiring information that quickly, however, comes at the expense of structural detail. Molecular dynamics simulations are also a valuable tool for studying protein configurations (see Physics Today, December 2013, page 13
Now Jaekyun Jeon, Robert Tycko, and coworkers at the National Institutes of Health in Bethesda, Maryland, have introduced a new way to track a protein’s folding.
1
Their experimental setup, shown in figure
Figure 1.

A rapid-mixing device starts and stops the protein-folding process with millisecond resolution. The mixer combines two pumped solutions, producing a high-velocity jet that freezes when it hits a liquid-nitrogen-cooled rotating copper plate. The frozen samples are subsequently analyzed using solid-state NMR. (Adapted from ref.

One moment in time
Melittin is a small protein—a peptide—with only 26 amino acids. At low pH, the peptides are linear chains, but in neutral to high pH, each peptide forms a bent helix. The helices form antiparallel dimers, which pair to make tetramers, melittin’s native configuration. 2 The entire transition happens in less than 10 ms, so whether those steps happen concurrently or sequentially has been hard to discern.
Tycko’s group developed a rapid mixer to change the solution’s pH and initiate the protein-folding process. It mixes two solutions in just 1.6 ms—not quite instantaneous on the protein-folding time scale, but fast enough to capture a narrow spread of folding times. Although it’s conceptually simple, the mixer is a critical part of their technique. “We’ve been working on these kinds of experiments for a while, and that was always one of our stumbling blocks,” says Tycko. The microfluidic devices the researchers tried previously were expensive and had to be discarded when they inevitably became clogged. In addition to being inexpensive, their small homemade mixer, made from standard chromatography fittings, is reliable and can be easily disassembled and rebuilt if it becomes clogged. And, importantly, it yields reproducible results.
The device will likely be useful for a wide range of future experiments because rapid mixing is a versatile way to trigger protein structural changes. The same mixer that Tycko’s group used to quickly change a solution’s pH could also be used for rapid dilution to alter a protein’s structure by changing the concentration of salt or of a denaturant such as urea or guanidine. Two interacting components could also be mixed to initiate complex formation.
To start melittin’s self-assembly process, the researchers mixed a low-pH solution of unfolded melittin with a high-pH buffer to produce a neutral solution. To stop the folding process, the mixer ejected the solution onto a liquid-nitrogen-cooled rotating copper plate that froze the solution—and the protein’s configuration—in less than 0.5 ms. The speed of the fluid leaving the mixer and the distance to the copper plate determined the protein’s structural evolution time. In experiments, the proteins were frozen after 2.2, 4.6, 9.4, and 29 ms. The initial low-pH solution provided measurements of the unfolded state, and a pre-mixed neutral solution represented the final folded configuration.
The researchers used carbon-13 isotope labeling to track the locations of three amino acids on each peptide: glycine-3, leucine-6, and isoleucine-20. Helix formation in melittin brings Gly3 and Leu6 close together, and dimer formation brings Leu6 and Ile20 close together, as illustrated in figure
Figure 2.

Melittin peptides fold and unfold in response to changing pH. At low pH they are extended chains (left), but at neutral to high pH they form ordered structures (right). Each chain forms a helix, the helices form antiparallel dimers, and the dimers pair into tetramers. The blue and white backbones indicate dimer pairs. Helix formation brings two labeled amino acids on a single peptide, glycine-3 (Gly3; purple) and leucine-6 (Leu6; green), closer together. When dimers form, the labeled isoleucine-20 (Ile20; orange) on one peptide gets close to the Leu6 on the other. (Adapted from ref.
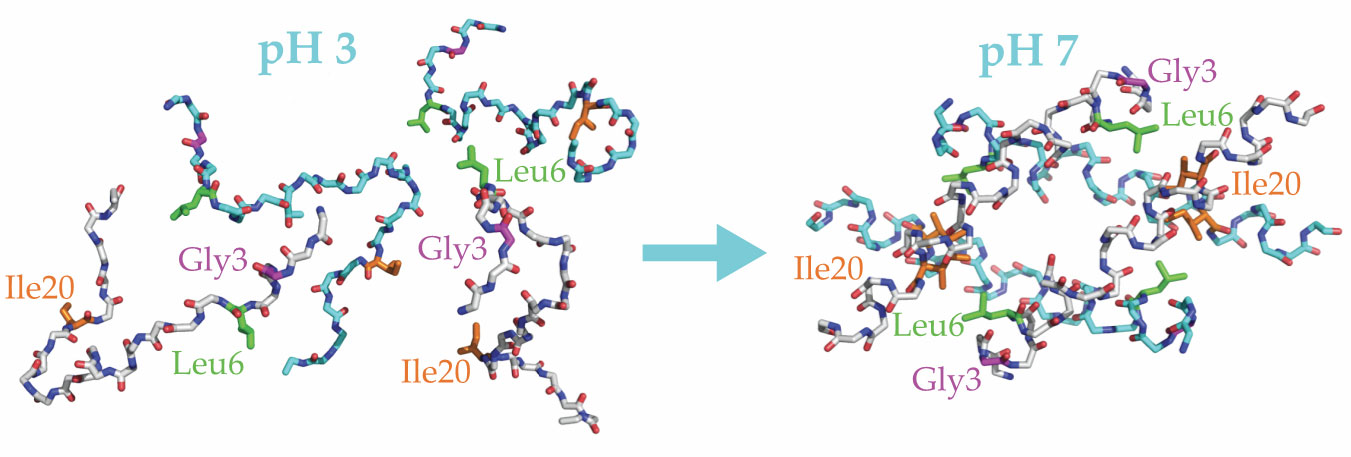
Freeze frame
Once they had a time series of frozen protein configurations, the researchers still faced the challenge of extracting structural information from their samples. Using NMR to discern the structures of biological molecules is a tried-and-true technique; Kurt Wüthrich received half of the 2002 Nobel Prize in Chemistry for developing such methods (see Physics Today, December 2002, page 19
Jeon, Tycko, and coworkers enhanced the ssNMR signal with dynamic nuclear polarization. DNP takes advantage of the fact that electrons are much easier to polarize than nuclei; the gyromagnetic ratio of an electron is about 2500 times that of a carbon nucleus. The electrons are irradiated with microwaves at the resonant frequency of their precession—263 GHz in the 9.4 T field from the lab’s homemade NMR setup—which flips some of the spins and reduces the electron polarization. When the electrons flip back to align with the field, they couple with the nuclear spins and, because they’re preferentially flipping in one direction, increase the nuclear polarization.
DNP is not a new technique—the underlying nuclear Overhauser effect was postulated and experimentally demonstrated 3 in lithium metal in 1953. But, as Tycko explains, it has had a renaissance in the past 10–15 years because of its newfound utility in biologically relevant experiments and improvements in microwave sources. 4 Applying DNP to time-resolved ssNMR made detailed protein measurements feasible: Previous experiments without DNP could not follow the evolution of intermediate structures. 5 The researchers achieved adequate NMR spectra in less than eight hours, whereas without DNP the same measurements would have taken a month or longer.
Come together
Jeon, Tycko, and coworkers used 2D NMR to track the development of helices and dimers in the melittin samples. They used a pulse sequence that transferred nuclear spin polarization between 13C atoms so that at the beginning of a measurement they observed the nuclear resonance from one atom, and at the end they observed that from another nearby atom. That’s what gives the technique its two dimensions—polarization transferring from one nucleus to another. The nuclei in different locations resonate at different frequencies, so the signal shows up as an off-diagonal peak, or crosspeak, in a two-dimensional NMR spectrum. The NMR pulse sequence was tailored such that nuclear polarization transfer between atoms in the labeled amino acids happened only when they were close enough (see Physics Today, October 2016, page 19
Figure
Figure 3.

Helix and dimer formation increase the transfer of nuclear spin polarization between labeled amino acids glycine-3 (Gly3), leucine-6 (Leu6), and isoleucine-20 (Ile20); the polarization transfer is quantified by the crosspeak volume ratio. The Gly3–Leu6 crosspeak, which indicates helix formation, and the Leu6–Ile20 crosspeak, which indicates dimer formation, both grow with the protein’s structural evolution time. The buildup times τb from exponential fits (dashed lines) are consistent with concurrent development of the two structures. (Adapted from ref.
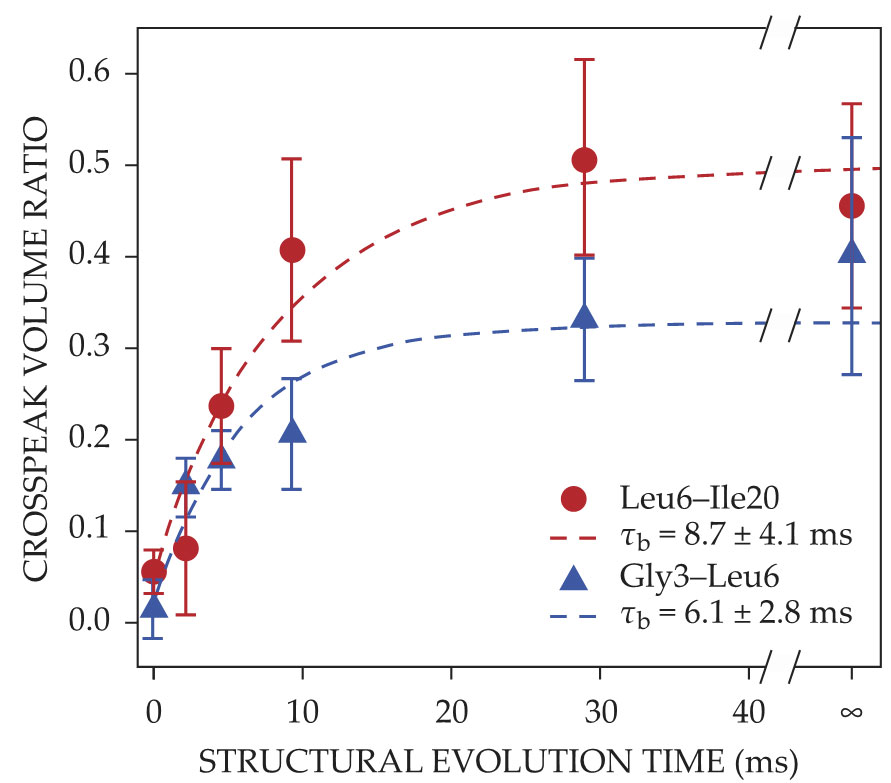
Time-resolved ssNMR was not used to track the protein’s full tetrameric structure. That experimental choice highlights one of the NMR technique’s limitations: The 2D spectra can be hard to interpret because the peaks are broad, particularly early on, when the protein is more disordered. Labeling more sites exacerbates the problem; the number of structural features that can be studied at once before the spectra become intractable is therefore limited. Although they could have done additional time-resolved ssNMR measurements with different labeled sites, the authors instead monitored tetramer formation using a less precise real-time fluorescence technique. Those measurements were consistent with tetramers forming along with the helices and dimers instead of the previously accepted sequential folding pathway. The unexpected result will inform theoretical models of protein folding dynamics.
Tycko sees time-resolved ssNMR as complementary to existing ways of studying protein folding. “There are a lot of other great techniques,” he says, “but they don’t give you the same kind of detailed molecular structural information that NMR can give you.” Now that they’ve demonstrated their method, the researchers are extending it to other protein-folding problems, such as complex formation by calmodulin (see Physics Today, May 2006, page 18
References
1. J. Jeon et al., Proc. Natl. Acad. Sci. USA 116, 16717 (2019). https://doi.org/10.1073/pnas.1908006116
2. T. C. Terwilliger, D. Eisenberg, J. Biol. Chem. 257, 6016 (1982).
3. A. W. Overhauser, Phys. Rev. 92, 411 (1953); https://doi.org/10.1103/PhysRev.92.411
T. R. Carver, C. P. Slichter, Phys. Rev. 92, 212 (1953). https://doi.org/10.1103/PhysRev.92.212.24. Q. Z. Ni et al., Acc. Chem. Res. 46, 1933 (2013). https://doi.org/10.1021/ar300348n
5. Y. Li et al., FEBS Lett. 377, 208 (1995); https://doi.org/10.1016/0014-5793(95)01338-5
K. N. Hu, W. M. Yau, R. Tycko, J. Am. Chem. Soc. 132, 24 (2010). https://doi.org/10.1021/ja908471n




