The cytoskeleton: I-beams of the cell
DOI: 10.1063/1.3326997
The cells that swim in the ocean , live in the ground, float in the air, and make up human tissue come in a wide variety of shapes, sizes, and stiffnesses that are integral to their biological functions and survival. In the early 1900s, biologists began to investigate the amazing deformations that some cells could undergo. They hypothesized that the mechanical properties of those cells are generated by a specific scaffolding of subcellular components, which was termed the cytoskeleton by Paul Wintrebert in 1931. A great deal of effort since then has produced a catalog of proteins that make up the cytoskeleton in eukaryotes—organisms, including humans and yeast, that compartmentalize their DNA into a nucleus. Those proteins self-assemble into an amazingly active material that is responsible for many of the animated motions associated with life.
The cytoskeleton of every eukaryotic cell is made up of two general classes of proteins: long polymers and cross-linkers. The three major kinds of polymers are microtubules, actin, and intermediate filaments. Each of those polymers has different geometrical, biochemical, and mechanical properties. Some cell types possess more specialized cytoskeletal filaments. Red blood cells, for instance, use a combination of fourfold and sixfold symmetric networks of the protein spectrin to achieve their remarkable plasticity—a necessity for cells that deform drastically as they circulate through small capillary openings. Proteins that link filaments make up the second integral component of the cyto-skeleton. A large number of such proteins exist, but they can be categorized as passive cross-linkers that bind filaments together or as active cross-linkers, often called molecular motors, that not only link two or more filaments but can actively convert chemical energy into mechanical work to apply forces to the filaments.
Active networks
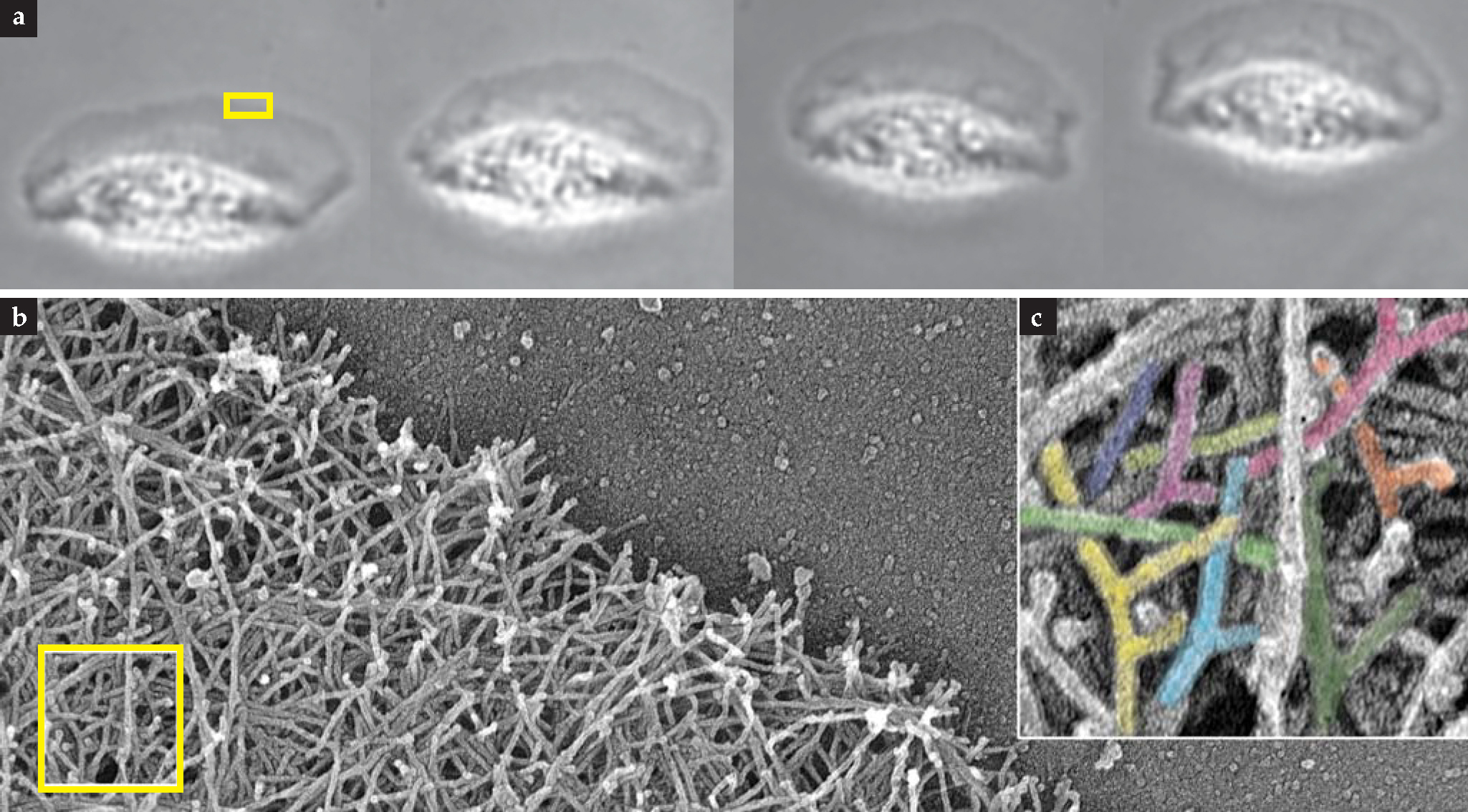
Cells constantly consume energy to grow, break down, and restructure their cytoskeletal networks. The mechanism by which many cells crawl on surfaces illustrates some key properties of an active cytoskeleton. The figure shows several views of a moving fish keratocyte, one of the fastest cellular crawlers. Fish keratocytes normally live on the outside of scales. When a scale is damaged, they are thought to migrate to the wound site to close it up and prevent infection.
A large flat region at the front of the cell, called a lamellipodium, is responsible for pulling along the bulky cell body at a rate of roughly one cell length per minute. To generate that fanlike region, tens of thousands of individual actin filaments form a branched network in which most filaments attach at 70° angles to other filaments. That structure is not static, however, and in a marvel of controlled self-assembly, freely diffusing actin proteins bind to the ends and sides of existing filaments, causing them to grow and branch only at the front of the cell. Because the actin network is attached to the underlying surface in discrete spots, growth at the front of the cell inches the cell forward. Constant growth, though, would quickly cause the cell—essentially a closed system — to run out of available actin protein. To solve that problem, tension generated near the rear of the cell by molecular motors, along with a specific set of localized enzymes and chemicals, detaches the network from the surface. The free actin filaments then break apart and decompose into unpolymeized actin protein that is transported to the front of the cell. The cycle of growth and destruction begins anew.
The gliding keratocyte can change direction in response to external signals from the environment. To do that, the cell must redefine which of its parts constitutes the front by relocating specific proteins there. That redefinition, combined with the local rules of network formation and growth, leads to a change in the overall direction of motion by increasing the rate of growth in one place and slowing it in another.
A stiffness measurement used by developing stem cells (cells that have not yet “decided” what type of tissue to become) to help in their decision making process reveals a different way in which the cytoskeleton is active. Molecular motors generate a force on the cytoskeleton that allows the cell to measure the elasticity of the underlying substrate. Those measurements, together with other signals, can induce a stem cell to become either a bone cell on stiff materials or a muscle cell on more compliant surfaces.
Self-assembled topologies
Nature has devised ways to build networks with different topologies—for example square, triangular, and radial networks; straight bundles; and the keratocyte’s network with its characteristic 70° branches. All of those networks possess a degree of inhomogeneity, but others are intrinsically random and disordered. How a cell creates a particular network is only partially understood, but self-assembly clearly plays a major role. Cells don’t use global blueprints for the design of cytoskeletal networks. Rather, the rules for network growth are set at the local level, built into the chemistry of each individual building block; self-assembly takes care of the rest. At the leading edge of the keratocyte, for example, activation of a protein complex called Arp2/3 produces the filament branches. Filament growth and branching together lead to long-range order. So reliable is this process that self-assembled cytoskeletal structures—radial arrays of microtubules, for instance—are used by cells as a means of ordering all other subcellular elements. Indeed, during cell division, the coordinated dynamics of two radial arrays of microtubules ensures that identical copies of each chromosome are aligned at the middle of the cell before they are whisked away to opposite edges.

Keratocyte cells use an active cytoskeleton to crawl on surfaces, (a) Successive frames from a movie show cell motion in the upward direction for one minute. The length of the cell is about 15 µm. (b) Electron microscopy (EM) of a portion of the cell’s front edge—the boxed region in panel a—shows the complex actin-filament network. (c) Increased magnification of part of that network (boxed region in panel b) reveals its characteristic Y-branch topology.
(EM images courtesy of Tatyana Svitkina.)
The complex geometry of a cytoskeletal network often leads to a counterintuitive, nonlinear deformation response to an applied force. On short time scales, the structural actin network that gives most cells their shape deforms readily when the force is large but is much stiffer when subjected to smaller forces. That allows a cell to experience large deformations when stressed but to return to its original shape afterward. When a force is applied over longer time scales, the networks tend to be viscous like maple syrup, a behavior that arises from the chemical dissociation and reassociation of the cross-linkers.
The cellular railroad system
In addition to generating structural support for the cell, cytoskeletal filaments serve as the tracks on which a number of molecular motors transport cargo. Most cytoskeletal polymers are polar and oriented, creating one-way roads on which the molecular motors travel. Moreover, each specific type of motor protein typically moves in one direction on only one kind of filament. By carefully controlling the activity of different motors, a cell can distribute molecules to specific locations in the cell. One of the most beautiful examples of that exquisite control is seen in amphibians and crustaceans that change color to blend in with their surroundings. Their real-time, on-demand camouflage is achieved by the dispersal or aggregation of pigment-containing granules to lighten or darken the skin. Granules are transported on both a radial microtubule array and a more randomly oriented actin network. By controlling the direction of microtubule-based transport and the chemical activity of the microtubule- and actin-based motors, the cell can either collect all the pigment at the cell center or disperse it uniformly throughout the cell.
New breakthroughs in our understanding of the cytoskeleton will require both experimental and theoretical effort. Experimentally, we currently lack the tools to perturb systems and visualize dynamics on the molecular scale. We therefore have only a limited ability to probe details of the changes in cytoskeletal topology in live cells as they go about their business. Future experiments that reveal the nanoscale dynamics of complex cytoskeletons should provide a wealth of new biological information. A theoretical understanding of the dynamics of those active, partially disordered, and inherently out-of-equilibrium systems will incorporate interesting problems from many areas of physics and engineering, including hydrodynamics, percolation theory, polymer physics, materials science, and statistical mechanics. The remarkable cytoskeleton promises to present a rich set of problems for the next generation of biological physicists to tackle.
References
1. J. Howard, Mechanics of Motor Proteins and the Cytoskeleton, Sinauer Associates, Sunderland, MA (2001).
2. D. Boal, Mechanics of the Cell, Cambridge U. Press, New York (2002).
3. T. D. Pollard, “The Cytoskeleton, Cellular Motility and the Reductionist Agenda,” Nature 422, 741 (2003).
4. K. E. Kasza et al.., “The Cell as a Material,” Curr. Opin. Cell Biol. 19, 101 (2007).
5. F. Jülicher et al.., “Active Behavior of the Cytoskeleton,” Phys. Rep. 449, 3 (2007).
More about the authors
Josh Shaevitz is an assistant professor in the department of physics and the Lewis-Sigler Institute for Integrative Genomics, and Simon Nørrelykke is a visiting fellow in the department of molecular biology, both at Princeton University in Princeton, New Jersey.
Joshua Shaevitz, 1 Lewis–Sigler Institute for Integrative Genomics, Princeton University, Princeton, New Jersey, US .
Simon Nørrelykke, 2 Princeton University, Princeton, New Jersey, US .




