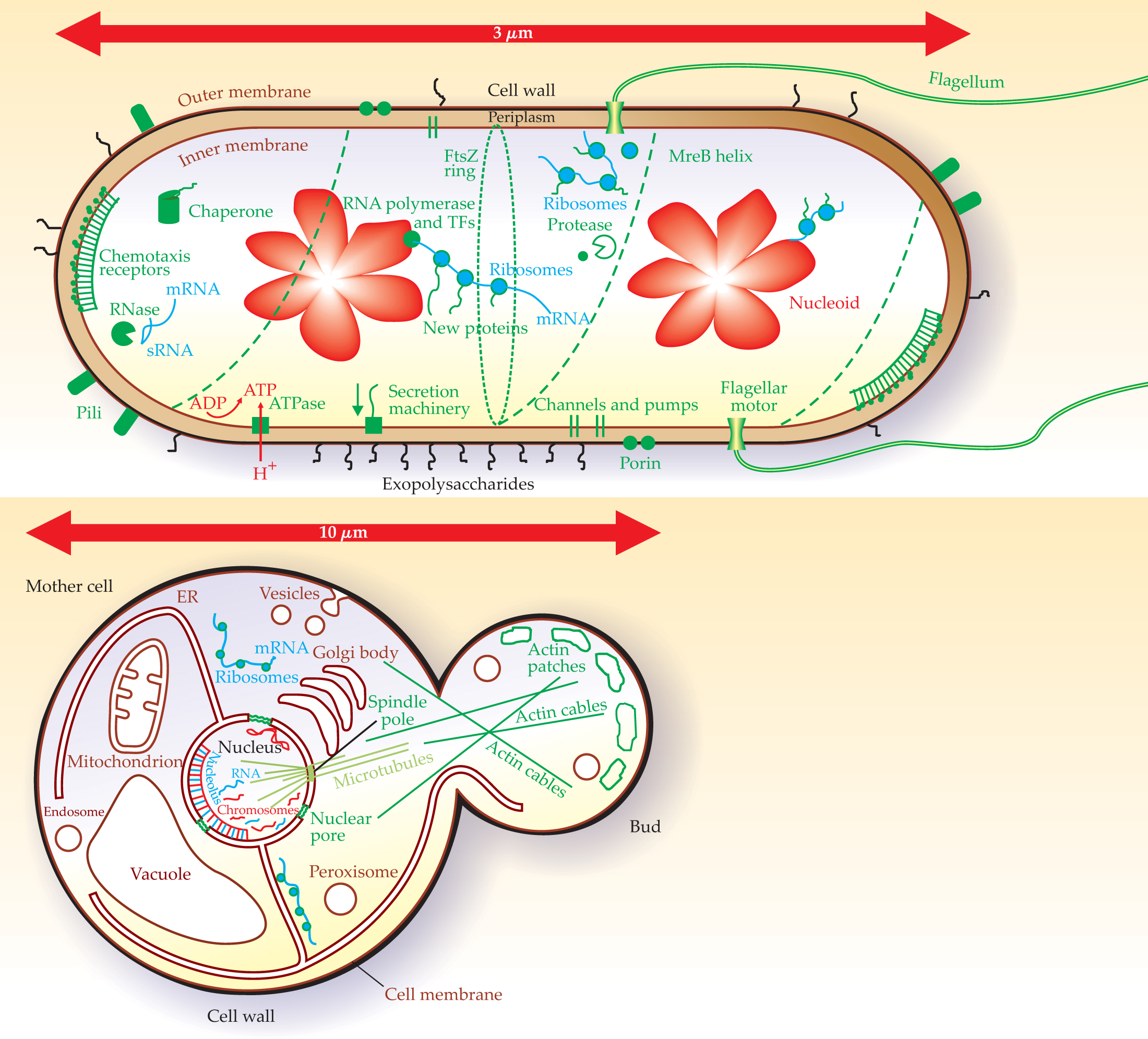
A glossary of cellular components
DOI: 10.1063/1.2364257
Among the most intensively studied organisms are the bacterium Escherichia coli and the yeast Saccharomyces cerevisiae. Biologists generally expect that a better understanding of those two model organisms will translate into a better understanding of all types of life. That expectation has been reinforced in recent years by results from genome-sequencing projects. Cells from bacterial to human all seem to use the same basic parts. That’s good for physicists, who generally don’t like to memorize a lot of names. However, even E. coli has about 4000 genes, each of which codes for a protein, and S. cerevisiae has about 6000. Biologists themselves can’t name all those genes, but they do know the names of the pathways and structures in which the proteins participate. To help physicists catch up, I’ve provided a brief glossary along with diagrams of E. coli and S. cerevisiae that indicate some of the most important structures.
General cellular structures
cell wall —A two-dimensional structure that provides strength. In E. coli, the cell wall lies between the inner and outer membrane and is made of peptidoglycan, a macromolecule composed of cross-linked peptide and sugar strands. In S. cerevisiae, the cell wall lies outside the membrane and is made primarily of polysaccharide chains.
channels and pumps —Proteins involved in, respectively, passive and active transport of small molecules through cell membranes.
chaperones —Proteins that help newly synthesized proteins to fold correctly or help in refolding misfolded proteins.
cytoplasm —The region inside the cell membrane, specifically inside the inner membrane of E. coli. In S. cerevisiae, the cytoplasm surrounds the nucleus and other organelles.
cytoskeleton —A framework of internal filamentous structures that provides for cell shape, transport, and, in eukaryotes, generation of forces via intracellular motor proteins.
DNA —A stable heteropolymer that forms a double helix by complementary base pairing. DNA contains the genetic information of a cell.
DNA polymerase —A complex of proteins that replicates DNA by making a complementary copy of a single DNA strand.
mRNA —Messenger RNA is transcribed on chromosomal DNA by RNA polymerase and is translated into protein on ribosomes. In bacteria, translation may begin during transcription, and a single mRNA transcript may contain genes encoding multiple proteins.
proteases —Proteins involved in the degradation of other proteins.
ribosome —An RNA-protein complex that translates mRNA into protein; the genetic code associates three consecutive mRNA bases (a codon) with a single amino acid of the protein.
RNA —A heteropolymer whose synthesis is directed by complementary base pairing with DNA templates.
RNA polymerase —A complex of proteins that transcribes the DNA template into RNA.
RNases —Proteins involved in the degradation of RNA.
secretion machinery —Proteins involved in intracellular transport, processing, and secretion of other specific proteins and molecules.
sRNA —Small RNA that can interact specifically with mRNA to regulate translation.
transcription factors (TFs) —Proteins that interact with DNA to activate or repress gene expression, that is, mRNA transcription by RNA polymerase.
Structures in E. coli
chemotaxis receptors —E. coli are capable of chemotaxis, that is, of swimming toward attractants and away from repellants. Cells use mixed arrays of five types of transmembrane chemotaxis receptors to detect temporal changes in external chemical concentrations. If conditions are getting better, cells continue to swim straight; if not, cells change direction.
flagellum —An extracellular structure, about 10 µm long, required for motility. An E. coli cell has roughly five flagella, each driven by its own rotary motor.
FtsZ —A protein that forms a ring on the cytoplasmic face of the inner membrane as the first step in cell division.
MreB —A protein that forms an extended spiral on the cytoplasmic face of the inner membrane. Disruption of MreB leads to cell shape changes, presumably because MreB helps organize the growth of new cell wall.
nucleoid —The condensed chromosome (DNA) of the cell. Prior to cell division, the chromosome is replicated, and the two chromosomes segregate to opposite halves of the cell and condense.
periplasm —The region between the inner and outer membranes.
pilus —Hair-like protein appendage involved in cell adhesion and DNA exchange.
porin —A ring of protein that forms a channel in the outer membrane.
Structures in S. cerevisiae
actin filament —Cytoskeletal polymer of actin protein about 10 nm in diameter. Cables made of actin filaments, in concert with motor proteins, are involved in cell motility, maintenance of cell shape, and transport of materials within cells.
chromosomes —Macromolecules of DNA that are stored in the nucleus and are organized by being wrapped around histone proteins.
endoplasmic reticulum (ER) —An organelle that extends from the nucleus and is involved in the modification and transport of proteins destined for the membrane or for secretion outside the cell.
endosome —A membrane-bound compartment in the cytoplasm involved in the degradation and recycling of membrane-bound proteins.
Golgi body —A membrane-bound organelle involved in protein modification and transport.
microtubule —Hollow tubule, about 25 nm in diameter, composed of protofilaments of tubulin proteins. Microtubules, which constitute part of the cytoskeleton, are involved in chromosome segregation during cell division and, with motor proteins, in cell motility, maintenance of cell shape, and transport.
mitochondrion —Organelle involved in synthesizing ATP (adenosine triphosphate), a molecule that stores chemical energy. Mitochondria derive from bacteria that became permanent residents of eukaryotic cells; about 100 are present in each yeast cell.
nuclear pore —A channel that allows transport of specific materials between the nucleus and the cytoplasm.
nucleolus —A region of the nucleus involved in the synthesis and processing of RNA to make ribosomes.
nucleus —An organelle in which chromosomal DNA is sequestered and replicated. RNA synthesized in the nucleus is exported as mRNA and other kinds of RNA. In yeast, the nucleus is approximately 1 µm in diameter and contains about 5 mm of DNA.
peroxisome —An organelle involved in lipid metabolism and protection of the cell from oxidation.
spindle pole —The organizing center for microtubules. The spindle pole is anchored in the nuclear envelope.
vacuole —An organelle involved in the storage and turnover of cytoplasmic substances.
vesicle —A small, membrane-bound compartment used for storage and transport.

Cellular structures. These schematic images of Escherichia coli (top) and Saccharomyces cerevisiae, or “budding yeast” (bottom), have been color coded and indicate DNA (red); RNA (blue); proteins (green), which, in actuality, densely pack the internal spaces; and lipids (brown). The E. coli bacterium is shown prior to cell division, after its chromosome (nucleoid) has been replicated. E. coli, which can divide every 30 minutes, is one of many bacteria found in the human digestive tract, but also survives well in external environments. S. cerevisiae, shown prior to chromosome replication, also has a long association with humanity—as an essential part of baking and fermenting. As with most eukaryotic cells including human cells, yeast cells are larger than bacterial cells and contain internal membrane-bound organelles such as the nucleus.
References
‣ B. Alberts et al., Molecular Biology of the Cell, 4th ed., Garland Science, New York (2002).
More about the authors
Ned Wingreen is a professor of molecular biology at Princeton University in Princeton, New Jersey.
Ned S. Wingreen, Princeton University, Princeton, New Jersey, US .




