Superresolution microscopy reveals chromosomes’ smallest structure
DOI: 10.1063/PT.3.2765
To fit in a cell nucleus, a strand of DNA must be folded to a size orders of magnitude smaller than its stretched length. In the first step of that compaction, the DNA winds around 10-nm protein clusters to form units called nucleosomes; the whole DNA–protein complex is known as chromatin. Cell biology textbooks typically illustrate chromatin’s structure as in figure 1a: with the nucleosomes evenly spaced along the DNA strand, like pearls on a string, and then arranged into a regular, compact 30-nm fiber.

Figure 1. Chromatin, the DNA–protein complex that makes up chromosomes, contains 10-nm units called nucleosomes. (a) Textbooks show the nucleosomes evenly spaced on the DNA strand, and then regularly packed into a 30-nm fiber. (b) New results show that, in fact, the nucleosomes are heterogeneously grouped into clusters, or “clutches.”
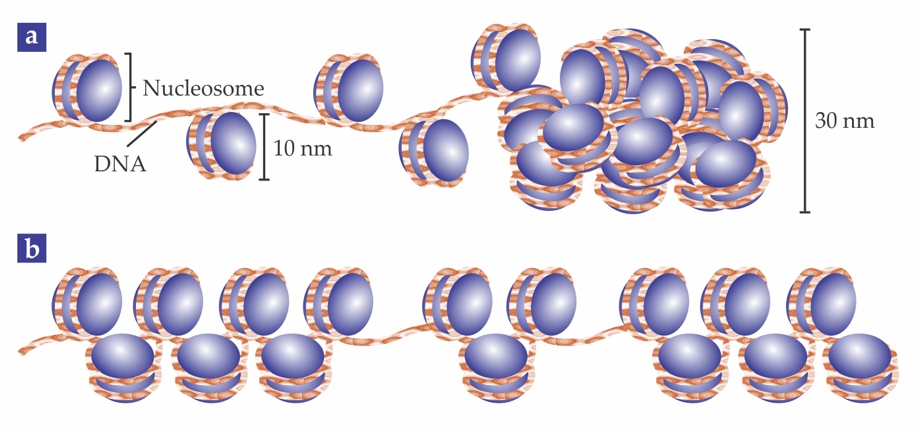
The evidence for that tidy picture is spotty—and, some have recently argued, wrong. But the traditional tools for probing cells have trouble with structures tens of nanometers large: Conventional optical microscopy doesn’t have the resolution, and electron microscopy and x-ray techniques lack the chemical specificity to distinguish the nucleosome proteins from other molecules.
Now Melike Lakadamyali, Maria Pia Cosma, and their colleagues at the Institute of Photonic Sciences (ICFO) and the Center for Genomic Regulation (CRG), both in Barcelona, Spain, are using superresolution microscopy to get a closer look at the structure of chromatin. 1 What they’ve found so far argues against the textbook view: Rather than being evenly spaced, nucleosomes are grouped into clusters, which the researchers call “clutches,” as shown in figure 1b. Furthermore, the average number of nucleosomes per clutch differs between stem cells and ordinary, or somatic, cells. Though they don’t yet know whether the clutch size is a cause or a consequence of the cell type, the researchers hope their work will lead to new insight into how stem cells differentiate into somatic cells—and how somatic cells can be “reprogrammed” into stem cells.
Open chromatin
Somatic cells contain all the necessary genetic information to perform any function in the body, but most of those roles are off-limits to them. Skin cells, for example, don’t spontaneously turn into liver cells. Embryonic stem cells, on the other hand, can divide to produce all cell types, so they’re prized for their potential use in cell-based regenerative therapies: the growth of new tissues to replace damaged ones.
Stem cells can be harvested from embryos, but that approach is fraught with both ethical controversy and practical limitations. A breakthrough came in 2007, when Shinya Yamanaka and his group at Kyoto University showed that they could reprogram human somatic cells into stem cells by introducing, via a virus, a few carefully selected extra genes. 2 (For his work, Yamanaka was awarded half of the 2012 Nobel Prize in Physiology or Medicine.) In the initial experiments, less than 0.1% of the somatic cells were converted to stem cells. Researchers have since improved the efficiency of the process by several means, including treatment with small-molecule drugs such as trichostatin A (TSA). 3
Cosma, a biologist at the CRG, was interested in understanding the mechanisms of that transformation process, particularly with regard to the chromatin structure. “It’s been said that TSA works by opening up the chromatin,” she says. “But nobody’s really sure what that means.” When she learned about superresolution microscopy as a promising new way to probe biological systems on an unprecedented length scale, she was inspired to get in touch with Lakadamyali, a physicist who was working on the technique at the ICFO across town.
Counting clutches
The Barcelona researchers used a superresolution method called stochastic optical reconstruction microscopy, or STORM, first presented in 2006 by Xiaowei Zhuang and her group at Harvard University.
4
(For a similar, simultaneously developed method, described in Physics Today, December 2014, page 18

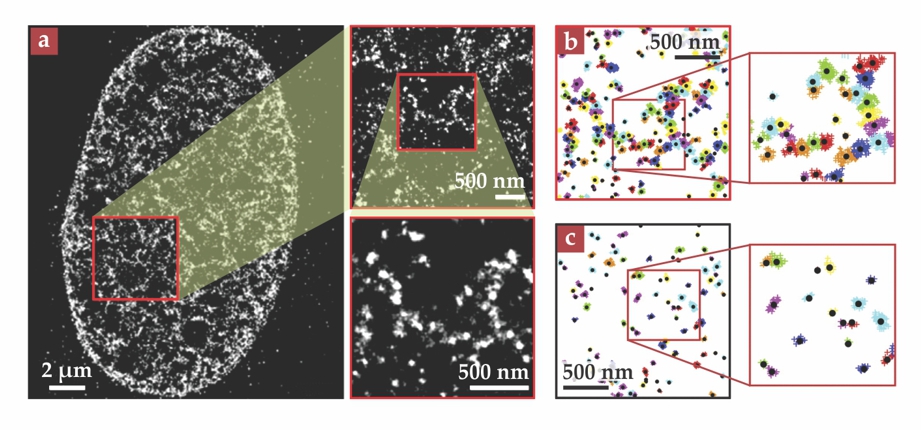
Figure 2. (a) Superresolution images show the heterogeneous arrangement of nucleosomes in a human somatic cell. (b) In data from the same cell, the bright spots are subdivided into nucleosome clutches, shown in different colors. (c) Smaller clutches are present in a somatic cell treated with trichostatin A, a drug that modifies chromatin structure and facilitates the reprogramming of somatic cells into stem cells. (Adapted from ref.
In STORM, as in conventional fluorescence microscopy, fluorescent molecules, or fluorophores, are attached to structures of interest. The fluorophores are optically excited with a laser, and the emitted light is captured with a camera. Each fluorescing molecule appears as a diffuse blob hundreds of nanometers wide, with that size governed by the wavelength of the emitted light, not the size of the molecule. But the centers of those blobs—the molecules’ actual positions—can be located with much greater precision, provided the blobs don’t overlap. Zhuang and company devised a system of fluorophores that could be switched repeatedly between fluorescent and nonfluorescent states. They could then switch off all but a few randomly chosen fluorophores, measure their positions with subwavelength precision, and repeat the process to build up a composite image with 20-nm resolution.
Although that’s not quite good enough to resolve individual nucleosomes, it would, Cosma and Lakadamyali anticipated, allow a good look at their arrangement—including the 30-nm fiber, if it existed. But when they captured their first images of somatic cell nuclei, they didn’t see continuous fibers at all. Instead, the images were blotchy and heterogeneous, with a lot of dark space, as shown in figure 2a. When they zoomed in as far as they could, they found the blotches to be composed of discrete bright spots tens of nanometers across.
At first, the researchers weren’t sure what to make of their images. But sophisticated numerical analysis ultimately showed that the blotches could be further subdivided into nanodomains, shown in different colors in figure 2b, each representing a clutch of nucleosomes that were close in space. They couldn’t directly count the number of nucleosomes in each clutch, but they could count the number of times a nucleosome from the clutch showed up in one of the component images that made up the STORM composite. Calibrating with images of structures containing known numbers of nucleosomes, they found that their somatic cells had a median of eight nucleosomes per clutch.
In stem cells, the researchers again saw evidence of clutches, but with median sizes between two and four. Somatic cells treated with TSA had similarly small clutches, as shown in figure 2c. And neural precursor cells—at an intermediate stage between stem cells and fully differentiated nerve cells—had clutches with a median of six nucleosomes.
Labeling the nucleosome proteins with fluorescent dyes required fixing the cells—that is, killing them—with an alcohol solution. To check whether their results were affected by that, or by any other aspect of their sample preparation, the researchers performed a series of control experiments, including one with living cells genetically modified to produce nucleosomes with fluorescent proteins already attached. None of the controls revealed anything different: The clutch structure showed up in all of them.
What’s next
The biological function of the clutches is not yet known. But they do indicate—as Carlo Manzo, an author on the paper, puts it—that “heterogeneity is much more important than we previously thought,” and the Barcelona researchers are just beginning to understand the full extent to which that’s true. For example, the segments of nucleosome-free DNA that connect the clutches are invisible in the STORM images, so the researchers don’t yet know just how the clutches arrange into larger structures. And they haven’t yet attempted to find out whether the clutch size varies between genes or between chromosomes.
The researchers plan to further explore how chromatin structure changes as stem cells differentiate and somatic cells are reprogrammed—by getting images from key points during the processes, or even by watching them in real time. They’re also interested in whether there’s any structural difference between chromatin in healthy somatic cells and in cancer cells.
“Superresolution microscopy is still quite a new field,” says Lakadamyali, “and the emphasis so far has been on technique development and proof-of-concept experiments. Only recently has there been a shift toward looking at real biological problems, and it’s very exciting.”
References
1. M. A. Ricci et al., Cell 160, 1145 (2015). https://doi.org/10.1016/j.cell.2015.01.054
2. K. Takahashi et al., Cell 131, 861 (2007). https://doi.org/10.1016/j.cell.2007.11.019
3. D. Huangfu et al., Nat. Biotechnol. 26, 795 (2008). https://doi.org/10.1038/nbt1418
4. M. J. Rust, M. Bates, X. Zhuang, Nat. Methods 3, 793 (2006).https://doi.org/10.1038/nmeth929
More about the authors
Johanna L. Miller, jmiller@aip.org




