Designer proteins fit like a glove
DOI: 10.1063/pt.ufgy.nwfg
The relationship between protein sequence and structure isn’t as much of a mystery as it once was. In late 2020, researchers from DeepMind in London turned heads with their AlphaFold2 model, which uses artificial intelligence to predict the structures of natural proteins with stunning accuracy. (See Physics Today, October 2021, page 14
Now researchers led by William DeGrado (University of California, San Francisco), his former postdoc Nicholas Polizzi (now on the faculty at Harvard University), and his current postdoc Lei Lu have unveiled a way to design proteins from scratch so that the proteins not only bind to a specified target molecule but do so with predictable binding energy. So-called de novo–designed proteins, made from amino-acid sequences that nature never exploited, are nothing new. But in most cases, the computed structures need to be experimentally refined with several rounds of mutation and screening before they’re fit for purpose. Polizzi and DeGrado’s proteins are notable exceptions: At least much of the time, they work on the first try.
The work builds on an underlying approach Polizzi and DeGrado developed several years ago, breaking down the protein–molecule binding problem into pieces called van der Mers. That’s not an obscure Dutch surname but rather a portmanteau of “van der Waals” and “rotamer.” Rotamers are floppy parts of amino acids; although they can, in principle, adopt enormously many conformations, only a handful of possibilities ever show up in the real proteins in the Protein Data Bank. By limiting themselves to just the conformations that nature tends to favor, protein designers can radically simplify their computational searches.

Starting from a few standard backbone structures, such as the alpha helices shown in gray, protein designers have extraordinary leeway to tune a protein’s chemical properties. Here, the amino acid side chains shown in purple and light blue are chosen to bind to a target molecule, shown mostly in pink. (Image adapted from L. Lu et al., Science 384, 106, 2024.)
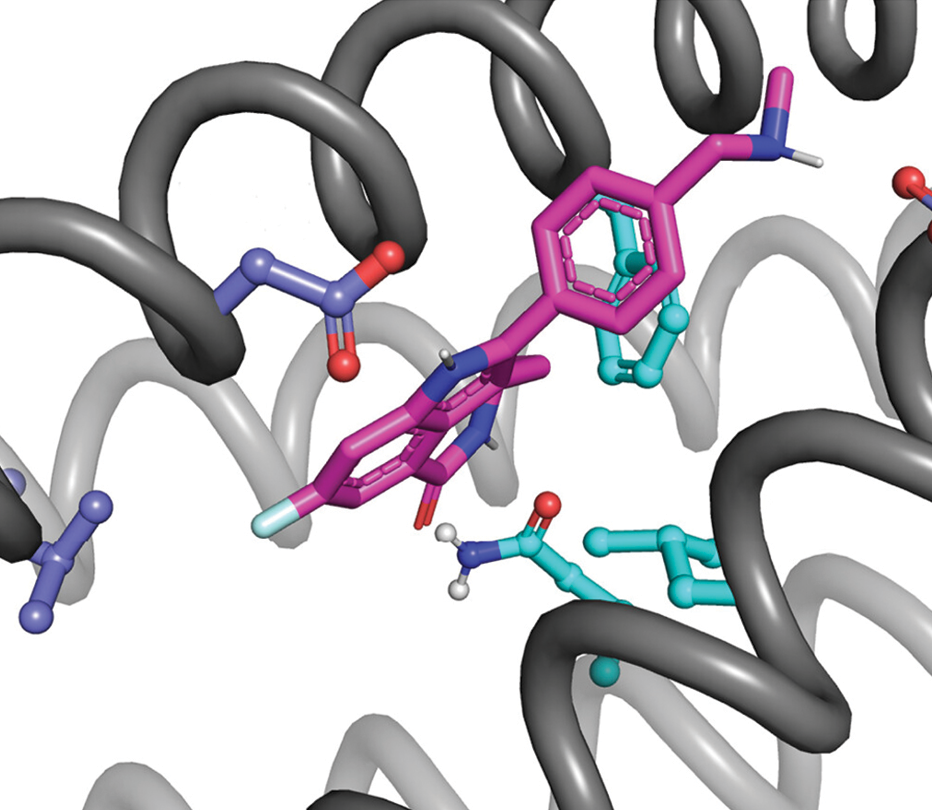
As Polizzi and DeGrado discovered, the same strategy works for the van der Waals interaction between amino acids and other molecules: Given an amino acid and the nearest bit of the target molecule, the atoms only ever arrange themselves in a few discrete ways. By sifting through the ways that the protein–molecule pieces can be packed into a known protein backbone—such as the four-helix structure shown in the figure—the researchers create a de novo protein fitted to the target molecule. With judicious choices of amino acids, they can make the binding as strong or as gentle as they like. And they can keep it specific: The protein binds to the target molecule, but not to any others.
The problem the researchers tackled is the inverse of conventional drug design. Given a naturally occurring protein, drug designers want to identify a molecule that binds to it, perhaps to stop it from performing some harmful function in the body. So why design a protein to bind to a given molecule? One possible application is to create antidotes for drugs—to neutralize them and stop their effects. Another is as a first step toward designing artificial enzymes: Before an enzyme can catalyze a reaction, it first needs to bind to the reactant molecule. (L. Lu et al., Science 384, 106, 2024
More about the authors
Johanna L. Miller, jmiller@aip.org




