Charpentier, Doudna win chemistry Nobel for development of CRISPR-Cas genome editing
DOI: 10.1063/PT.3.4630
Designer babies. Reviving the woolly mammoth. With prospective uses that sound more like science fiction than science fact, it’s no wonder that CRISPR-Cas—the versatile biomolecular tool that gives researchers the power to make precise changes to cells’ genetic codes—has captured the popular imagination.
But beyond the hype—and the thorny ethical questions—CRISPR-Cas is making waves throughout the biosciences. Inherited diseases whose genetic basis is well understood, such as cystic fibrosis and sickle cell anemia, could become routinely treatable. Agricultural crops could be more easily engineered with sturdier stems, better drought tolerance, and other desirable features. And across the basic-research community, CRISPR-Cas is revolutionizing what scientists can do with cells and DNA, even in contexts seemingly far removed from genome modification.
“I think of it as the equivalent of equipping your car with a GPS system,” says Jie Xiao of Johns Hopkins University. “All you have to do is program in the coordinates and you can go anywhere in the genome that you want. You can bring lots of tools with you, and when you get there, you can do anything.”
Emmanuelle Charpentier HALLBAUER & FIORETTI, BRAUNSCHWEIG, GERMANY Jennifer Doudna UNIVERSITY OF CALIFORNIA, BERKELEY



The discovery of CRISPR-Cas as a tool stems from a 2012 experiment by the groups of Emmanuelle Charpentier (then at Umeå University in Sweden, now at the Max Planck Unit for the Science of Pathogens in Berlin) and Jennifer Doudna (University of California, Berkeley). 1 Now the Royal Swedish Academy of Sciences has chosen Charpentier and Doudna for this year’s Nobel Prize in Chemistry.
From bacteria to biotechnology
Although commonly called a gene editor, CRISPR-Cas is more precisely considered a gene cutter. In its canonical form, it seeks out a specific sequence of DNA and snips both strands of the double helix. To introduce the desired edits—to add, delete, or modify base pairs—researchers harness a cell’s existing pathways for repairing its own DNA. The methods for doing so were already part of the pre-CRISPR-Cas genetic engineering toolbox.
The toolbox also contained two other gene cutters: zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). Both those classes of proteins target DNA just like CRISPR-Cas does. The DNA sequence they target, however, requires an amino-acid sequence in the protein to recognize and interact with it. Getting that sequence just right—and getting the protein to fold into the right conformation—is a difficult and laborious process.
The beauty of CRISPR-Cas is that the target DNA sequence is simply the complement of an RNA guide sequence held by the Cas (short for CRISPR-associated) protein. Targeting a new gene is as easy as swapping out one piece of RNA for another.
The name CRISPR itself, which stands for “clustered regularly interspaced short palindromic repeats,” has nothing to do with genome editing—and everything to do with where CRISPR-Cas systems were discovered in nature: as part of a microbial immune system. Viruses attack bacteria by injecting their genetic material into the single-celled organisms and commandeering their cellular machinery to make more viruses. To fend off those attacks, bacteria use CRISPR-Cas to cut the viral DNA and render it harmless.
To know which strands to cut, the bacteria store in their genomes short segments of DNA from every virus that they (or their ancestors) have ever encountered. Those viral snippets are separated from one another by a series of identical, symmetrical DNA sequences—the CRISPR.
The unusual repeating motifs in bacterial genomes were first observed in 1987, and their immune function was elucidated in the mid to late 2000s. A small but vibrant research field sprang up to identify and classify the various Cas proteins present in different bacterial species and to figure out how they worked in conjunction with other molecules to protect bacteria from viruses. The nascent discipline attracted both Charpentier, a microbiologist, and Doudna, a biochemist.
In their landmark 2012 discovery, Charpentier, Doudna, and their research groups built a simplified CRISPR-Cas system, outside of its native bacterium, that they could program with RNA of their choice to target and cut any DNA. 1 They chose the protein Cas9, from the bacterium Streptococcus pyogenes, for its all-in-one function: Unlike some bacterial species, which employ teams of several different Cas proteins to recognize and cut the target DNA, S. pyogenes uses just one.
A key insight was that the guide RNA, shown in red in figure
Figure 1.

Adapted from a bacterial immune system, the CRISPR-Cas9 gene editor consists of the Cas9 protein (light blue) and a strand of guide RNA (red) that contains a 20-nucleotide guide sequence. The complex seeks out and binds to the complementary DNA sequence; two active sites (HNH and RuvC, purple) then cleave the DNA. (Adapted from ref.
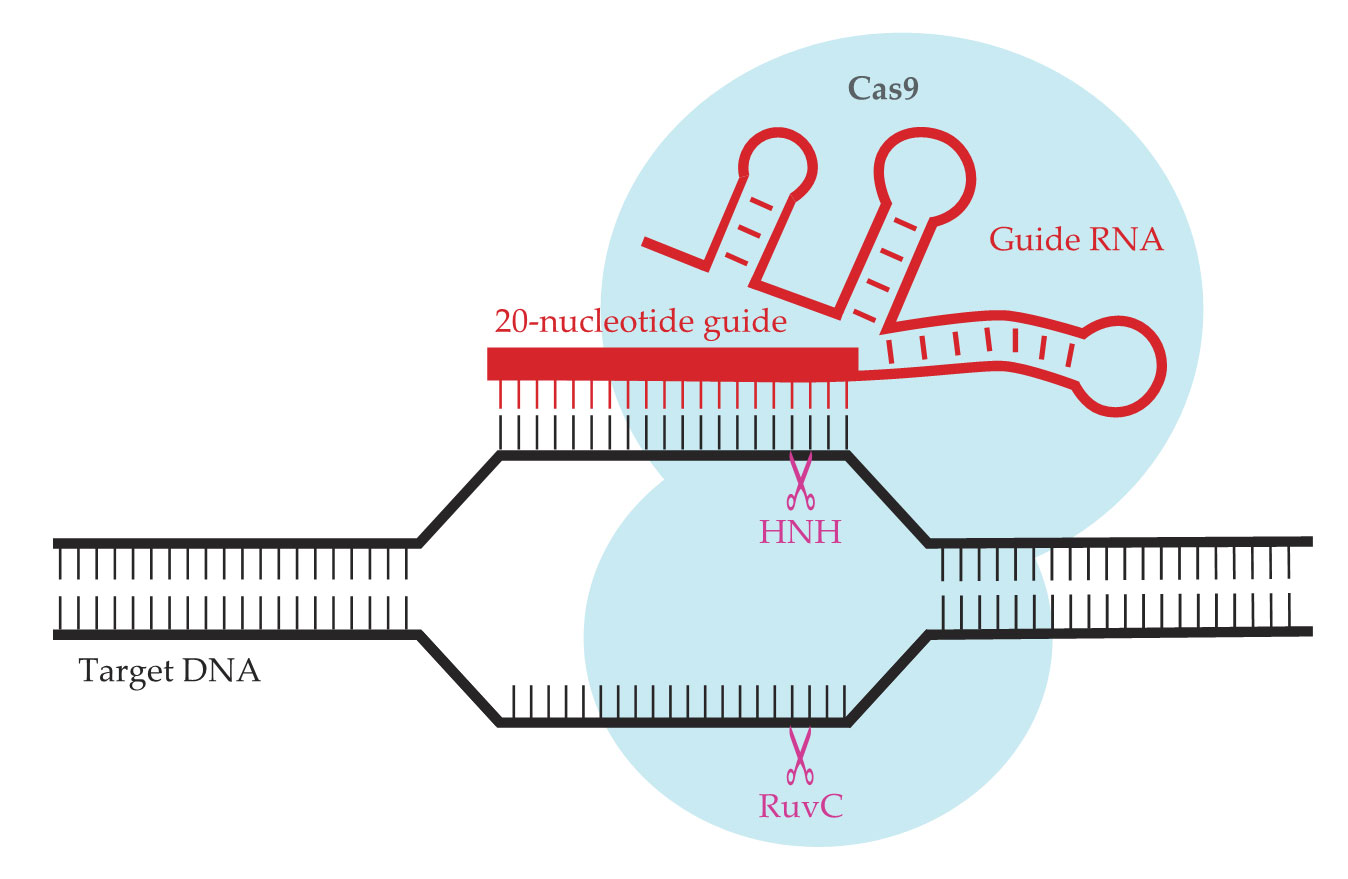
When Charpentier and Doudna demonstrated CRISPR-Cas9’s programmable DNA-cutting activity in vitro, it still wasn’t obvious that the system would work on the genomes of higher organisms. Plants and animals have a lot more DNA than either viruses or bacteria do, and they package it in dense, compact chromosomes inside a cell nucleus. As it turned out, however, none of those potential obstacles is enough to keep CRISPR-Cas9 from finding its target: Within just a few months of Charpentier and Doudna’s publication, three other groups showed that the gene cutter can be made to work in human cells. 2
On target
Viruses mutate over time. For CRISPR-equipped bacteria to mount an effective defense, their immune systems should be able to recognize viral DNA that isn’t quite identical to the viruses they’ve encountered before. And indeed, wild-type CRISPR-Cas systems can cleave DNA that differs from their target sequence in two or three places.
But for editing human genes, it’s important to get an exact match. The 3 billion base pairs in the human genome (counting just 1 of each of the 23 pairs of chromosomes) is small compared with the 1 trillion possible 20-nucleotide Cas9 target sequences. It’s unlikely that a target sequence will occur in two or more unrelated places in the genome. But when CRISPR-Cas9 also targets near-miss sequences, the chances of it cutting the wrong gene are much greater.
Guided by theoretical and experimental studies of how CRISPR-Cas9 works, researchers have introduced mutations into the Cas9 structure to increase its specificity and limit the so-called off-target effects. (See the article by Giulia Palermo, Clarisse G. Ricci, and J. Andrew McCammon, Physics Today, April 2019, page 30
But there’s a delicate balance to be struck. CRISPR-Cas9 consumes no ATP or any other molecular source of energy. The energy it needs to unzip the target DNA must be supplied by the energy released when the complex binds to the target sequence, so if that interaction is made too weak, the system won’t work. Ahmet Yildiz of the University of California, Berkeley, notes that although several versions of Cas9 with enhanced specificity have been developed, “it’s still a challenge to avoid off-target effects without compromising the efficiency of on-target editing.”
The gene cutter that doesn’t cut
Tweaking Cas9’s amino-acid sequence can not only refine the protein’s activity but also change it. As figure
The cuts and repairs induced by regular Cas9 are permanent, and that’s too heavy a hand for some experiments on gene function. But when dCas9 binds, temporarily, to its target gene, it blocks the cell’s transcription machinery from accessing that gene—in effect, switching it off. The ability to turn genes on and off at will lets researchers investigate how genes interact with one another to form circuits and how cells control which genes they express and which ones they don’t. Almost every cell in the human body has all the same DNA, but the proteins in a muscle cell differ from those in a nerve cell. The capacity of cells to regulate their gene-expression profiles is essential to the development of complex multicellular organisms.
And dCas9 can do more to control gene activity besides physically blocking its target gene. Before sending dCas9 into the cell, researchers can affix to it one of the cell’s own DNA-processing proteins, such as an activator or a repressor, so that when dCas9 binds to its target sequence, it boosts or restricts the expression of the gene next door. “Previous methods for doing this were not so straightforward,” says Yildiz, “but with dCas9, we can block or control genes, change the gene expression profile, and switch from gene to gene without much trouble.”
Furthermore, dCas9 can be decorated with fluorescent tags that enable researchers to image the location of the target sequence inside the nucleus—and ultimately the spatial arrangement of the whole genome. It’s long been known that chromosomes’ folded conformations are intricately hierarchical. But the leading method for probing them, called Hi-C, has its limitations.
In a Hi-C experiment, the cell is flooded with chemical links that bind DNA strands that are close in space; sequencing the DNA on either side of the links reveals which genes on which chromosomes are positioned together inside the cell. (See Physics Today, December 2009, page 19
Figure 2.

Just a few of CRISPR-Cas’s many applications. (a) Genes tagged with fluorescently labeled dCas9 (green) are imaged at different stages of mitosis. (b) The cytoskeletal protein vimentin (green) in a living cell is made fluorescent by modifying its gene with CRISPR-Cas9. (The protein tubulin, purple, is stained with a fluorescent dye.) (c) Target DNA from a virus is detected with Cas12a, which goes on to cut the DNA tethers holding reporter molecules together. Once cut, the reporter molecules become fluorescent. The detection scheme can be used to test for COVID-19. (Panel a adapted from ref.
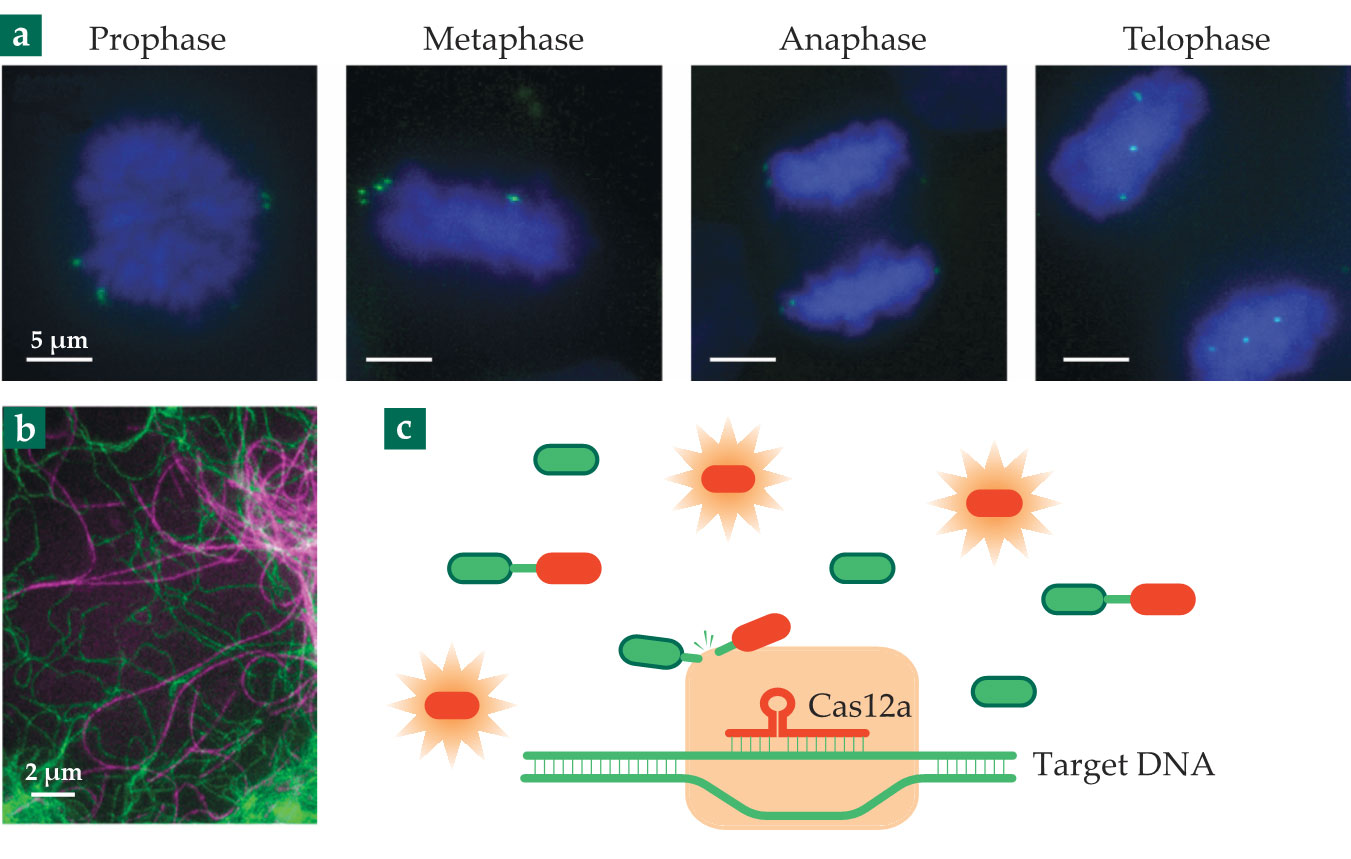
Bringing cells into view
Imaging applications of CRISPR-Cas systems extend beyond looking at the genome itself. Fluorescence imaging can uncover hidden structures in all parts of the cell—and with superresolution techniques, it can do so in spectacular detail. (See Physics Today, December 2014, page 18
For chemically fixed (that is, dead) cells, components can be labeled by staining the cell with fluorescent dye molecules bound to antibodies. The antibodies seek out and attach themselves to the protein molecules of interest, whose locations in the cell can then be imaged.
But dead cells aren’t suitable for studying cellular dynamics. For imaging live ones, the usual technique is to force the cells to synthesize so-called fusion proteins: the protein of interest bound to either a fluorescent protein (see Physics Today, December 2008, page 20
The plasmid-endowed cell has two versions of the protein gene: the one it carries naturally and the fusion gene on the plasmid. To maximize their chances of seeing the protein fluoresce, researchers typically induce the cell to overexpress the plasmid gene—that is, to make so many copies of the fusion protein that it overwhelms the cell’s natural production of the untagged protein.
That overexpression, however, throws the cell’s protein populations out of balance, and researchers can’t be sure that the cell they’re studying is representative of the cell’s natural state. “If you have too much of the fusion protein and all its natural localizations in the cell are occupied, the excess proteins might go somewhere else,” explains Stefan Jakobs of the Max Planck Institute for Biophysical Chemistry. “That may result in a wrong conclusion about the proteins’ subcellular localization.”
Genome editing solves that problem: If researchers can modify the cell’s existing gene to include the fusion component, they can study the fusion protein expressed in natural amounts. They’ve been doing just that, with ZFNs and TALENs, before CRISPR-Cas technology came along, but CRISPR-Cas’s programmability has sped the process, and it’s now routine to obtain images such as the one in figure
A world of possibilities
Much of the CRISPR-Cas research so far has used modifications of the Cas9 protein from S. pyogenes. But the microbial world is full of other Cas proteins, and even other versions of Cas9. Cas proteins are classified by their function and their evolutionary relationships, but even ones with the same name can have considerable structural differences. The Cas9 from Staphylococcus aureus, for example, has two cutting sites that resemble those of the S. pyogenes Cas9, but it’s almost 25% smaller.
The diminutive size is important for some medical applications. “When it comes to treating blood diseases, we can extract bone marrow cells from the patient, do genome editing in a petri dish, then reintroduce them into the patient,” says Dipali Sashital of Iowa State University. “But editing things like liver cells is a lot more challenging, and it requires a more complicated delivery mechanism.” Benign viruses, stripped of their genetic material, can be loaded with the ingredients of a CRISPR-Cas system and deliver them to cells in the body, but only if the payload is not too large. The S. aureus Cas9 is small enough to fit, but the S. pyogenes Cas9 is not.
Despite the diversity of Cas9 proteins, they’re an overall minority in nature. Many more microbial species employ so-called type I systems, which have a markedly different behavior: A complex of several proteins—Cas5, Cas7, and Cas8—recognizes the target sequence, then recruits another protein, Cas3, which not only cuts the target DNA but demolishes a large part of it.
“Evolutionarily, that probably makes more sense,” says Sashital. In its function in a bacterial immune system, a CRISPR-Cas system needs to render viral DNA unusable. The sledgehammer of Cas3 is at least as effective in that role as is the scalpel of Cas9. Although type I systems aren’t well suited for precise gene editing, their capacity to make large genome deletions has its uses—for example, in experiments that seek to identify which genes are essential to a microorganism’s survival. 4
One of CRISPR-Cas’s most promising practical applications comes from two relatively new additions to the Cas toolkit, Cas12a and Cas13. Like Cas9, both are all-in-one proteins that detect the target genetic material—DNA for Cas12a, RNA for Cas13—and attack it, and both make precise cuts in their target sequences. But they don’t stop there. Binding to the target sequence turns both proteins into indiscriminate genome-cutting machines, cleaving any other DNA or RNA that happens to be around, regardless of its sequence.
Because they can cut many strands of DNA or RNA with just one detection of the target sequence, Cas12a and Cas13 can function as biosensors. The concept is shown in figure
In particular, that scheme can be used to detect viruses by their characteristic genetic material. Developed in 2018, the technique has been quickly pursued by companies that include Mammoth Biosciences in California (cofounded by Doudna) and Sherlock Biosciences in Massachusetts. With the outbreak of the COVID-19 pandemic, both companies have adapted their platforms to test for the SARS-CoV-2 virus. “Our test was the first FDA-approved CRISPR product,” says James Collins, a cofounder of Sherlock Biosciences, “and we’ve contracted with Integrated DNA Technologies to produce a million tests a week.”
The DNA strands in the reporter molecules are not part of the code of life. Rather, they serve as structural elements—a polymer tether that Cas12a happens to be able to cleave. Repurposed DNA has been explored for many other materials science applications, either by itself or integrated with other materials (see, for example, Physics Today, April 2012, page 20
Any number of DNA-based materials can now be made to respond to CRISPR-Cas systems in potentially useful ways. Before the pandemic, Collins and his group demonstrated a smart hydrogel made of DNA and other polymers. When Cas12a detected its target DNA sequence in the environment, it would cleave the DNA in the hydrogel to either release a cargo molecule or change the material’s mechanical or electrical properties. “This line of research is just getting started,” says Collins. “But a lot of researchers are wondering, ‘What else can we use this for?’”
References
1. M. Jinek et al., Science 337, 816 (2012). https://doi.org/10.1126/science.1225829
2. L. Cong et al., Science 339, 819 (2013); https://doi.org/10.1126/science.1231143
P. Mali et al., Science 339, 823 (2013); https://doi.org/10.1126/science.1232033
S. W. Cho et al., Nat. Biotechnol. 31, 230 (2013). https://doi.org/10.1038/nbt.25073. B. P. Kleinstiver et al., Nature 529, 490 (2016); https://doi.org/10.1038/nature16526
I. M. Slaymaker et al., Science 351, 84 (2016). https://doi.org/10.1126/science.aad52274. See, for example, B. Csörgő et al., Nat. Methods (2020), doi:https://doi.org/10.1038/s41592-020-00980-w
5. F. Jiang, J. A. Doudna, Annu. Rev. Biophys. 46, 505 (2017). https://doi.org/10.1146/annurev-biophys-062215-010822
6. B. Chen et al., Cell 155, 1479 (2013). https://doi.org/10.1016/j.cell.2013.12.001
7. A. N. Butkevich et al., ACS Chem. Biol. 13, 475 (2018). https://doi.org/10.1021/acschembio.7b00616
More about the authors
Johanna L. Miller, jmiller@aip.org




