Nuclear Magnetic Resonance Spotlights Atomic Actors in Enzyme Dynamics
DOI: 10.1063/1.1485566
Without enzymes, life—from aardvarks to zucchini—would stop dead. Fortunately, thanks to enzymes’ catalytic power, biochemical reactions run briskly at temperatures low enough that life’s molecular building blocks don’t boil to bits. Like other bio-molecules, enzymes rely on their form and flexibility for their function. Only if an enzyme can adopt the right shape can it latch onto its customary substrate and lower the activation energy of the reaction it catalyzes.
Enzymologists suspect that enzymes change shape throughout the catalytic process—from the initial binding, through the chemical transformation, to the final release. But tracking those changes of shape is difficult. Crystallography, the structural biologist’s workhorse, is limited by its nature to the study of rigid assemblies that flex only slightly. Its practitioners can take snapshots of the few points in the enzymatic cycle that crystallize, but can’t shoot a movie of the whole cycle.
But now, a team led by Dorothee Kern of Brandeis University in Waltham, Massachusetts, has used nuclear magnetic resonance (NMR) to provide the first atomic-resolution look at the dynamics of enzyme action. 1 Her study, which paired an enzyme called cyclophilin A (CypA) with a simplified version of its natural substrate, does not identify all the atomic participants or even how they move. But it does furnish enough detailed information to build a plausible model of how CypA does its job.
Flexible backbone
Like most enzymes, CypA is a protein. As such, it’s made up of a characteristic sequence of amino acids strung together with peptide bonds that form when one acid’s NH2 end hooks up with another acid’s COOH end. Together, the peptide bonds compose the protein’s backbone.
As a starting point, Kern assumed that any important conformational changes that CypA undergoes would show up in its peptide backbone rather than in its side chains, which, being attached to the backbone by single bonds, flop about too much to form the basis of a reliable, repeatable reaction scheme. The three-dimensional structure of CypA is already known, thanks to x-ray crystallography. To map how the structure changes, Kern and her colleagues turned to NMR.
NMR is sensitive to a nuclear spin’s chemical environment because the electrons that swarm around the nucleus alter its magnetic environment in a predictable way. This “chemical shift” in the nucleus’s NMR frequency is characteristic of an atom’s chemically bound neighbors, whose valence electrons it shares. If a nucleus moves to a different magnetic neighborhood—if the molecule it inhabits changes shape—then its chemical shift changes too.
Except for hydrogen, the atoms normally found in proteins have no nuclear spin. The lack of spins is actually an advantage for studying molecular motion because a CypA sample can be created in which all the molecules have a nucleus with nonzero spin at the same individual location. Kern—or, rather, her specially cultured bacteria—created 160 different versions of CypA (one for each of the enzyme’s peptide bonds) in which the more commonplace spin-0 nitrogen-14 is swapped in the peptide bond for spin-1/2 nitrogen-15.
Kern, along with postdoc Elan Eisenmesser and grad student Daryl Bosco, placed sample solutions of enzyme and substrate in a strong magnetic field, which pulls the spins into line with the field’s axis (labeled z, by convention). A brief magnetic pulse then flips the spins momentarily onto the xy-plane. As the spins try to realign with the z-axis, they gradually lose their coherence with one another. The rate at which they lose coherence, known as the transverse relaxation rate R 2, turns out to be especially well suited to probe conformational changes that take place on the microsecond-to-millisecond time-scales characteristic of enzyme action.
But NMR by itself can’t map structural changes. Also needed is a way of ascertaining what reaction state the enzyme is in. At a given instant, a CypA molecule could be unattached to a substrate molecule; it could be in the midst of binding to the substrate; it could be catalyzing the substrate’s conversion to the product molecule; or it could be releasing itself from the product. At the same instant, the countless other CypA molecules in the sample could be in any of those states.
Sorting out what’s going on in this molecular melee would be impossible but for a simple and clever tactic that Kern devised. Her idea relies on CypA’s membership in the class of enzymes known as isomerases.
An isomerase catalyzes the transformation of a molecule into a different version of itself—termed an isomer—that has the same chemical formula but a different structure. CypA works only on proteins that contain the amino acid proline. Of the various types of isomeric transformation, CypA performs the simplest: the rotation of part of protein about one of the bonds connected to proline. With the enzyme’s help, one rotational configuration (the cis isomer) transforms into the other (the trans isomer)—and vice versa.
This reversibility is the key. In a lab solution of CypA and substrate, the conversion between the two isomers keeps turning over indefinitely. Kern realized that the enzymatic cycle of the sample as a whole could be tipped toward binding or catalysis by controlling the relative concentrations of enzyme and substrate.
To see how the idea works, think of the enzyme molecules as the hosts of a party and the substrate molecules as their guests. When an enzyme host meets and grasps the hand of an apprehensive substrate guest, the two bind. The host catalyzes the greeting by shaking the guest’s hand and uttering words of welcome. To complete the greeting, the host releases the guest’s hand, transforming the guest’s initial anxiety into a feeling of ease.
In the lab, each experimental run—one per peptide bond—begins with a fixed concentration of enzyme and a much lower concentration of substrate. Without much substrate to work on, enzymes spend more time binding than they do catalyzing. As more substrate is added, catalysis begins to predominate. And when the enzyme is saturated with substrate, catalysis occurs at its maximum rate. At that point in the enzyme–substrate party, everybody is busy shaking hands.
The changes in configuration that occur during enzymatic action are measurable because they change the transverse relaxation rate R 2 in a predictable way. If a 15N nucleus takes part in the configuration change, its chemical shift will fluctuate between two values. As a result, its spin will lose coherence at a faster rate and R 2 will increase. For the boost in R 2 to be significant enough to measure, the time the molecule spends changing configuration must be roughly reciprocal to the difference in chemical shifts. Fortunately, that happens to be the case. At the 14-tesla fields used in the Brandeis experiments, the chemical shifts tend to be about 1000 Hz, with differences of about 10 Hz.
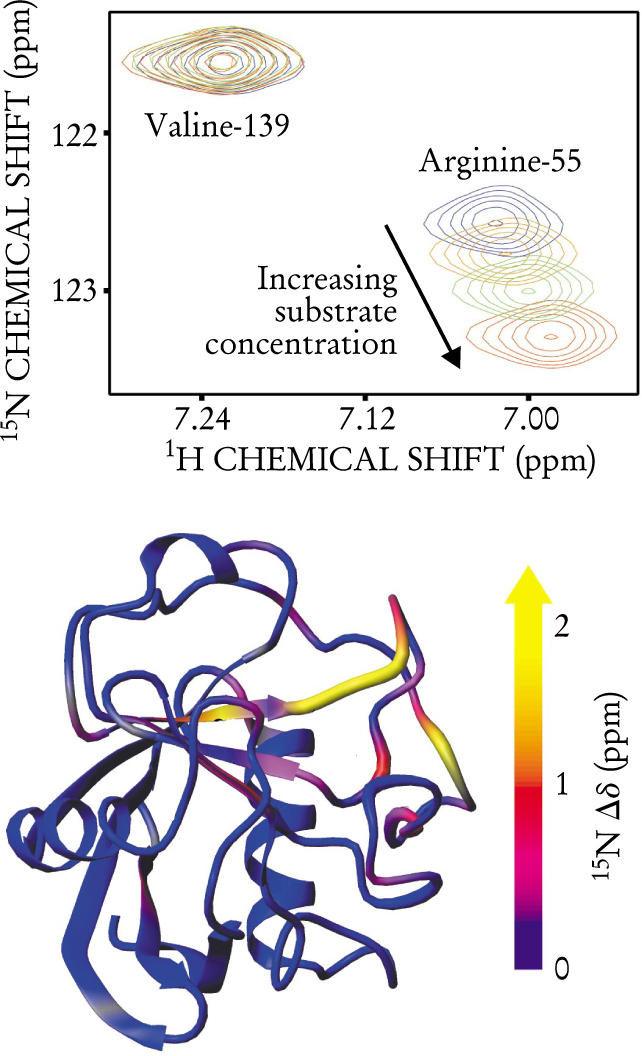
Figure 1 shows the results of looking at 15N nuclei in the peptide bonds attached to two of CypA’s amino acid components, arginine-55 and Valine-139. (The numbers refer to the amino acid’s position in the protein, counting from the protein’s N terminus.) As the substrate concentration increases, the magnetic environment around valine-139 remains constant through the enzymatic cycle. That’s not the case for arginine-55, whose chemical shifts change systematically with increased substrate concentration. Evidently, arginine-55 takes part in enzymatic action, whereas valine-139 does not. Other peptide bonds in CypA move during the enzymatic cycle, as indicated in the bottom of figure 1. Most, however, are bystanders.

Only some of CypA’s amino acids take part in enzyme action. The top plot shows chemical shifts for the hydrogen and nitrogen-15 nuclei in two peptide bonds: in valine-139 and in arginine-55. (Chemical shifts are measured in parts per million with respect to the resonance of the free nucleus.) As substrate concentration increases, the chemical shifts of arginine-55 change systematically. Valine-139, however, shows no change in chemical shift. The bottom diagram shows which parts of CypA move during the enzymatic cycle. It was created by subtracting the chemical shifts of inactive enzyme from those of active enzyme and mapping them onto the enzyme’s structure.
(Adapted from ref. 1.)
But Kern’s team went beyond mapping the enzyme’s active regions. Kern and her colleagues could also distinguish the subregions that bind the substrate from those that catalyze it. To do so, she first assumed that CypA’s enzymatic cycle lacks complicating intermediate steps and involves only the steps of binding, catalysis, and release. If that’s the case, then for an amino acid that participates only in catalysis, R 2 will keep increasing as more and more substrate is added, or—in party terms—as more and more guests arrive. But for an amino acid component that participates only in binding and release, the biggest boost to R 2 occurs when free and bound enzyme are both present in significant amounts, or—back to the party—when the number of guests hasn’t reached the point that hosts have difficulty finding unoccupied guests to greet.
Applying this approach to each of the amino acids, Kern found that only one, arginine-55, turns out be the catalyzer, confirming a result that Lynne Zydowsky (now at Renovis in San Francisco) and her colleagues had derived 10 years ago using site-specific mutagenesis. Eight amino acids, including arginine-55, play the role of binders.
To check the assumption of a three-step process, Kern’s collaborator Mikael Akke of Lund University in Sweden used a simple physical model to calculate how R 2 varies as a function of substrate concentration. His model nicely matched the NMR data for the amino acids that move during the enzymatic cycle.
The NMR data distinguish between binding and catalysis, but can’t reveal the difference between cis and trans binding because both are present in the lab samples at the same time. Fortunately, in 1996, Yingdong Zhao and Hengming Ke of the University of North Carolina at Chapel Hill derived the crystal structure of CypA bound to the cis—and only the cis—isomer of a peptide. Armed with Zhao and Ke’s structure, Kern could deduce by elimination which of the binding amino acids move when they lock onto the trans isomer.
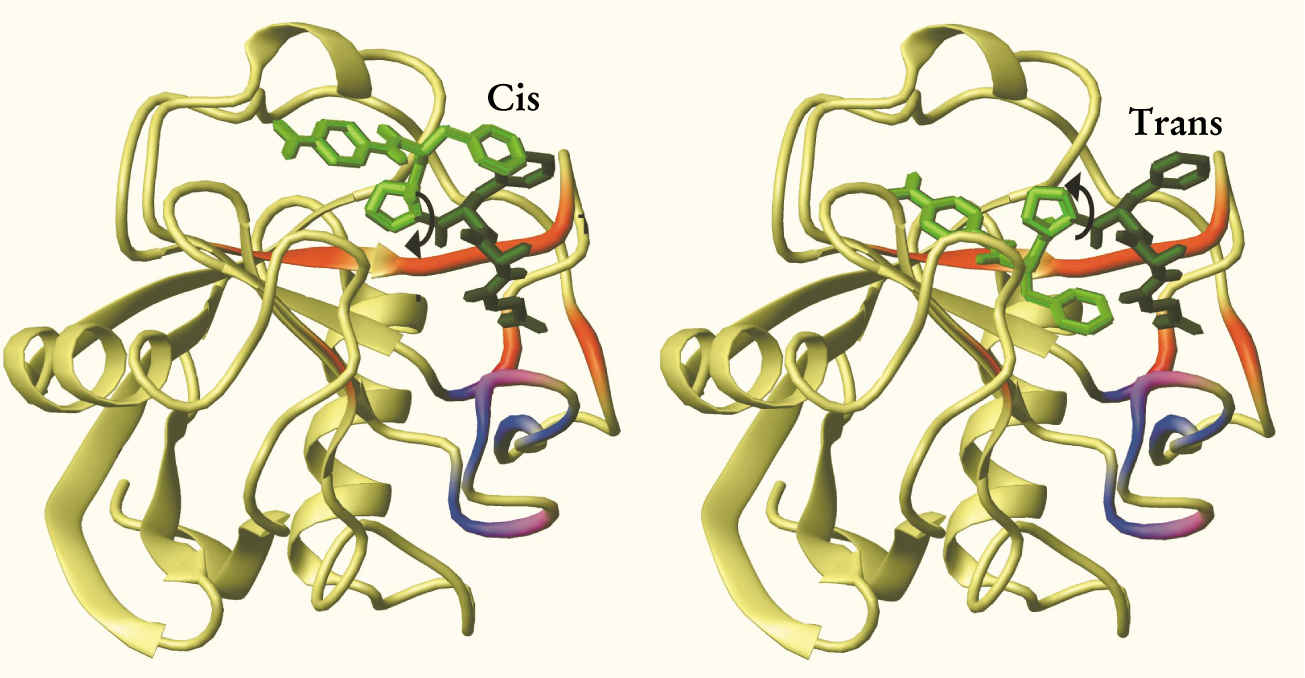
With that last piece of the puzzle in place, she and her colleagues could come up with the reaction pathway shown in figure 2. In the case of cisto-trans isomerization, the first step is that the cis isomer sticks to six of the eight amino acids identified as binders. Next, arginine-55 catalyzes the isomerization by weakening the stiff single bond between the proline and its neighboring amino acid. With one part of the substrate still stuck to the enzyme, the other part detaches from the enzyme and rotates 180° around the weakened bond, forming the trans isomer. The rotated end sticks momentarily to the remaining two amino acid binders before the enzyme releases the isomerized substrate. The trans-to-cis isomerization works the same way, but in reverse.

The cis-to-trans isomerization by CypA (shown mostly in yellow-green) of a model protein substrate (medium and dark green). Throughout catalysis, part of the cis isomer (dark green) remains bound to the enzyme. The other part (medium green) rotates by 180° about the proline bond to form the trans isomer. The parts of the enzyme that participate in the enzymatic cycle are shown in red. (The parts shown in blue and purple move even when no substrate is present; most likely they have no role in enzymatic activity.)
(Adapted from ref. 1.)
Transplants, HIV
The very property that makes CypA amenable for Kern’s NMR investigation—that the enzyme catalyzes a reversible isomerization—makes it hard to identify CypA’s overall purpose in cellular activity because substrate and product are so similar. Indeed, when it was first discovered in 1984, CypA was named not for its enzymatic activity—which was unknown at the time—but for its ability to bind to and disable cyclosporin A, a drug that suppresses human immune systems in organ transplants.
CypA was identified as an enzyme in 1989, when two groups, 2 one from Tonen’s corporate R&D lab in Saitama, Japan, and one from the Max Planck Research Unit for Enzymology of Protein Folding in Halle, Germany, proved that CypA is identical to a prolyl isomerase, which, like CypA, had been discovered in 1984. The story didn’t end there. CypA and its fellow cyclophilins have turned up in all sorts of places in all sorts of organisms, including the capsid of the HIV-1 virus.
Now, evidence is emerging that CypA’s fundamental role is tied to its enzymatic function—even when it appears merely to bind to other proteins. Kern and her collaborators recently used NMR to test whether CypA isomerizes HIV capsid protein. 3 It does. The finding suggests that HIV-1’s deadly virulence could depend on CypA’s catalytic ability.
References
1. E. Z. Eisenmesser, D. A. Bosco, M. Akke, D. Kern, Science 295, 1520 (2002). https://doi.org/10.1126/science.1066176
2. N. Takahashi, T. Hayano, M. Suzuki, Nature 337, 473 (1989). https://doi.org/10.1038/337473a0
G. Fischer et al., Nature 337, 476 (1989). https://doi.org/10.1038/337476a03. D. A. Bosco et al., Proc. Natl. Acad. Sci. USA 99, 5247 (2002). https://doi.org/10.1073/pnas.082100499




