Visualizing DNA biomechanics
DOI: 10.1063/PT.3.1927
As nanoscale structures and devices become more and more sophisticated, understanding their behavior—mechanical, electrical, and other—becomes increasingly important. Biological nanostructures can offer particular promise but also unique challenges, given their fragility and the difficulties in probing them. Ulrich Lorenz and Ahmed Zewail now report using ultrafast electron microscopy (UEM) to directly visualize nanomechanical motions of DNA structures in space and time. Key to the technique is stroboscopic imaging: Exposing a sample to a series of coherent, precisely timed single-electron packets in an electron microscope builds up, over the course of several experimental runs, a real-time motion picture of the sample, with up to 1012 frames per second.
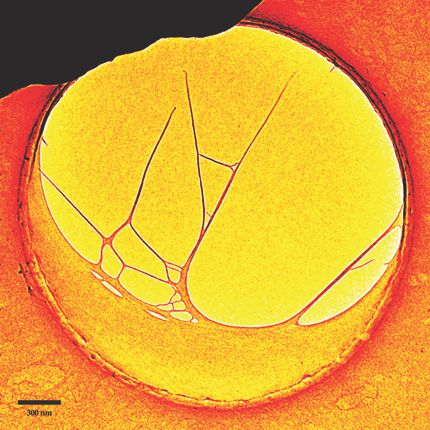
This micrograph shows a sample for one of the duo’s DNA movies. A drying aqueous solution of bacteriophage DNA formed a crescent-shaped sheet across the lower part of this 2.5-µm hole in a carbon substrate. On the left, thin filaments 20–30 nm in diameter had originally extended to the upper rim before the pair severed them with a tightly focused electron beam. Like one would strike a tuning fork, the researchers hit the carbon substrate with a picosecond laser pulse to induce oscillations in the free-standing DNA nanostructure. (Focusing the laser at the substrate protected the DNA from damage.) From the vibrations recorded by their UEM movie, the pair could extract such information as the normal modes, resonant frequencies, and Young’s moduli of various network parts. The approach, the two argue, is readily generalized to mechanics studies of other biological nanomaterials and networks. (U. J. Lorenz, A. H. Zewail, Proc. Natl. Acad. Sci. USA, in press.)
To submit candidate images for Back Scatter, visit http://contact.physicstoday.org