Newfound links close the loop on a gene regulatory network
DOI: 10.1063/PT.3.2131
Like humans, Escherichia coli require a carbon-rich diet to thrive. And like humans, the bacteria sometimes become set in their dietary ways. When glucose is readily available, for instance, E. coli stop producing enzymes used to metabolize alternative carbon sources such as lactose and maltose. Cambridge University biochemists Helen Epps and Ernest Gale, who observed a version of that peculiar response in the 1940s, dubbed it the glucose effect. Now known to be triggered by certain other sugars as well, the effect has come to be regarded more broadly as catabolite repression.
One of the earliest discovered examples of gene regulation, catabolite repression is widely thought to be mediated by the phosphotransferase system (PTS), a series of reactions that transport glucose and a handful of other sugars into the cell. The reactions also have the effect of inhibiting the synthesis of cyclic adenosine monophosphate (cAMP), a messenger molecule tasked with activating the synthesis of several sugar-processing enzymes—the same enzymes that go missing during catabolite repression.
The PTS–cAMP mechanism has become a textbook illustration of how cells use signaling pathways to adapt to different nutrient supplies. But as an explanation of catabolite repression, it isn’t entirely satisfying. A number of groups have observed catabolite repression triggered by sugars that aren’t transported by the PTS. In other cases, catabolite repression has been triggered not by a particular sugar but by a shortage of noncarbon ingredients such as nitrogen.
Now an international collaboration led by Terence Hwa (University of California, San Diego), Dalai Yan (Indiana University School of Medicine, Indianapolis), Peter Lenz (University of Marburg, Germany), and Yi-Ping Wang (Peking University, Beijing) has devised a model that can explain many of those apparent anomalies. 1 In the new model, cAMP remains the key player, but it takes on a new, expanded regulatory role.
“Nothing special”
The group’s model was inspired by an experiment carried out by Conghui You, a postdoc in Hwa’s lab. In the course of growing E. coli on more than a dozen different carbon sources, she discovered that concentrations of LacZ, one of the cAMP-activated enzymes affected by catabolite repression, seemed to obey a simple, generic trend: When plotted against growth rate, as shown in figure 1, it always fell on the same downward sloped line—regardless of the nutrient composition. The concentrations of half a dozen other cAMP-activated enzymes were observed to behave in the same way.

Figure 1. Escherichia coli’s experimentally measured growth rates and enzyme levels reveal a simple, seemingly universal trend: As growth rate falls, the concentration of certain carbon-metabolizing enzymes—including LacZ, plotted here—increases. Each data point represents a different carbon source. Growth rate is expressed as the inverse of the population doubling time, and enzyme concentration is expressed in terms of Miller units, where 1 Miller unit corresponds to roughly 1 enzyme molecule per cell. (Adapted from ref.
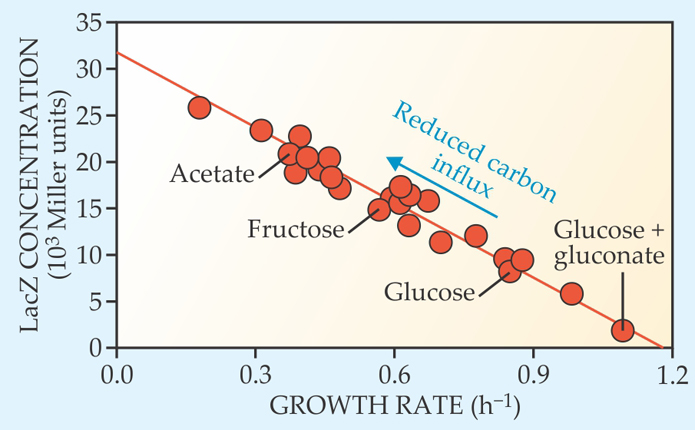
You’s microbes were grown under carbon-limited conditions, meaning that their growth rates were determined by their capacity to ingest and metabolize carbon. The implication, then, is that the more quickly a cell can metabolize carbon, the less cAMP it produces. Glucose is a readily metabolizable sugar, which helps explain the earlier studies linking it to low cAMP concentrations. But otherwise, explains Hwa, “it is nothing special.” Apparently, E. coli’s cAMP messengers respond to the sugar according to the same rules that govern their response to every other carbon source. So what are those rules?
The protein factory
Microbial growth requires the coordinated effort and interactions of thousands of genes and proteins. (See the article by Rob Phillips and Stephen Quake, Physics Today, May 2006, page 38
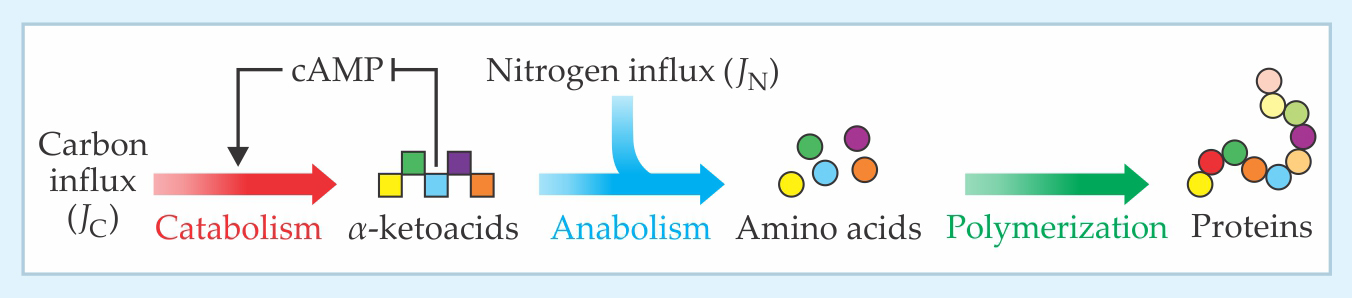
Hwa and his coworkers simplified the task by classifying E. coli’s biological machinery according to a coarse-grained division of labor: Catabolic proteins, including LacZ and other cAMP-activated enzymes, convert carbon-based nutrient molecules into α-ketoacids; anabolic proteins assimilate nitrogen and other essential ingredients used to convert α-ketoacids into amino acids; and ribosomal proteins polymerize the amino acids into new proteins.
How fast the cells grow depends on how efficiently they deploy their protein resources. A cell can increase the rate of carbon uptake by increasing the expression of genes encoding catabolic proteins, but that does little good if there’s insufficient nitrogen to convert the carbon into amino acids. Likewise, the mass deployment of ribosomal proteins would be wasteful if the levels of anabolic and catabolic proteins were too low to supply a steady stream of amino acids.
Hwa and his coworkers figured that E. coli might be using cAMP to maintain a balance between catabolic and anabolic fluxes in order to prevent bottlenecks in its amino-acid assembly line. That would explain the apparent negative correlation between cAMP and carbon influx: When carbon fluxes are low compared with nitrogen fluxes, it makes sense to deploy more catabolic proteins and fewer anabolic ones; when carbon fluxes are relatively high, the opposite strategy is advantageous.
Figure 2 illustrates how such a regulatory scheme might be implemented. The α-ketoacids serve as a natural indicator of how well the cell is balancing its catabolic and anabolic needs: They accumulate when the carbon influx JC outpaces the nitrogen influx JN and deplete when the imbalance is the other way around. If one assumes that α-ketoacids exert a negative feedback on the production of catabolic proteins—presumably by inhibiting the synthesis of cAMP—the resulting model yields a growth-rate trend much like that seen in figure 1.

Figure 2. In a coarse-grained model of Escherichia coli metabolism, catabolic proteins degrade carbohydrates into molecules known as α-ketoacids; anabolic proteins assimilate nitrogen and convert α-ketoacids into amino acids; and ribosomal proteins polymerize amino acids to build new proteins. The α-ketoacids accumulate or deplete depending on the relative carbon (JC) and nitrogen (JN) fluxes. A balance is maintained with the help of a negative feedback loop, in which α-ketoacids inhibit the production of cAMP, a messenger molecule that activates the synthesis of catabolic proteins. (Adapted from ref.
To test the theory, the researchers added various α-ketoacids directly into the nutrient supply of a growing E. coli culture. Almost instantly, the microbes stopped synthesizing LacZ and other cAMP-activated enzymes. The same was true for strains of E. coli that had been genetically modified to lack the PTS pathways previously thought to dictate cAMP levels. With in vitro experiments, the group was able to confirm that the α-ketoacids themselves—and not their biochemical byproducts—were inhibiting the synthesis of cAMP.
“This helps answer a lot of long-standing questions,” says molecular biologist Thomas Silhavy of Princeton University. Among them is the issue of how carbon catabolite repression can be induced by nitrogen depletion. In Hwa and coworkers’ model, the difference JC − JN determines the feedback signal, so a shortage of nitrogen has the same effect as an excess of carbon.
A fresh approach
In a way, Hwa and company have turned the standard approach to systems biology on its head. Instead of trying to piece together details of individual biomolecular interactions in order to predict the behavior of an entire organism, the researchers investigated the behavior of the entire organism to gain insights into poorly understood molecular-scale interactions.
“It’s not based on exotic experimental techniques that no one has ever seen or used before,” says Uwe Sauer, a systems biologist at ETH Zürich. “They’ve just used rigorous thinking to reveal a simple behavior that was lying under our noses the whole time.”
Sauer thinks the approach could work for a number of other physiological puzzles that continue to vex researchers in the field. One of them, the so-called Warburg effect, in which certain cells grow anaerobically even in oxygen-rich environments, already has Hwa’s attention. Says Hwa, “Our lab is working on that as we speak.”
References
1. C. You et al., Nature 500, 301 (2013).https://doi.org/10.1038/nature12446




