Neutron scattering for structural biology
DOI: 10.1063/PT.3.4498
The past two decades have seen explosive growth in research on structural molecular biology. High-throughput techniques for determining biological structures are yielding large amounts of information about atomic-, cellular-, and tissue-scale organization. Advances are driven by modern high-brilliance synchrotron sources, synchrotron-based full-field x-ray microscopy and tomography, free-electron lasers, and cryoelectron microscopy facilities. The scientific landscape is changing at a remarkable pace with increasing emphasis being placed on interdisciplinary and multi-technique approaches. Neutron scattering facilities around the globe are expanding their capabilities to provide unique and complementary insights about biological systems. 1
Neutron-based techniques have become increasingly powerful and sophisticated tools for structural biology research. Recent improvements in sources, instrumentation, and sample preparation have revolutionized the scope of biological neutron scattering. Although such methods have been used to probe macromolecules
2
since the 1960s, their application to the study of biological systems was limited because data acquisition was technically difficult and frustratingly slow. (For more about early work on biological neutron scattering, see the article by Peter Moore, Physics Today, January 1985, page 62
Neutrons around the world
There are two types of modern neutron beam sources. Continuous, or steady-state, sources produce neutrons by fission, whereas pulsed sources produce neutrons by spallation—the breakup of nuclei. The two types of sources have different yet complementary properties. Continuous sources use classical optical elements such as choppers, monochromators, and analyzers to condition neutron beams and parse scattering data. Pulsed sources have an inherent time structure that derives from their use of proton accelerators; that structure allows for time-of-flight measurements and offers the ability to discriminate between particle energies. Neutrons produced by both types of sources typically have energies similar to those associated with atomic and molecular motions. To study macromolecular systems under physiologically relevant conditions, researchers use cold neutrons with wavelengths from 40 Å to 2 Å and thermal neutrons with wavelengths less than 2 Å because both types probe biologically relevant length and time scales.
Neutron facilities that undertake structural biology research exist throughout the world (see figure
Figure 1.

Neutron facilities are found across the globe. Some, like the Institut Laue–Langevin, have been engaged in biological neutron research for decades. The European Spallation Source, on the other hand, is still under construction. Once operational, it will be the world’s most powerful spallation source. (Tetiana Chemerys/Alamy Stock Photo; adapted by Freddie Pagani using www.neutronsources.org/neutron-centres
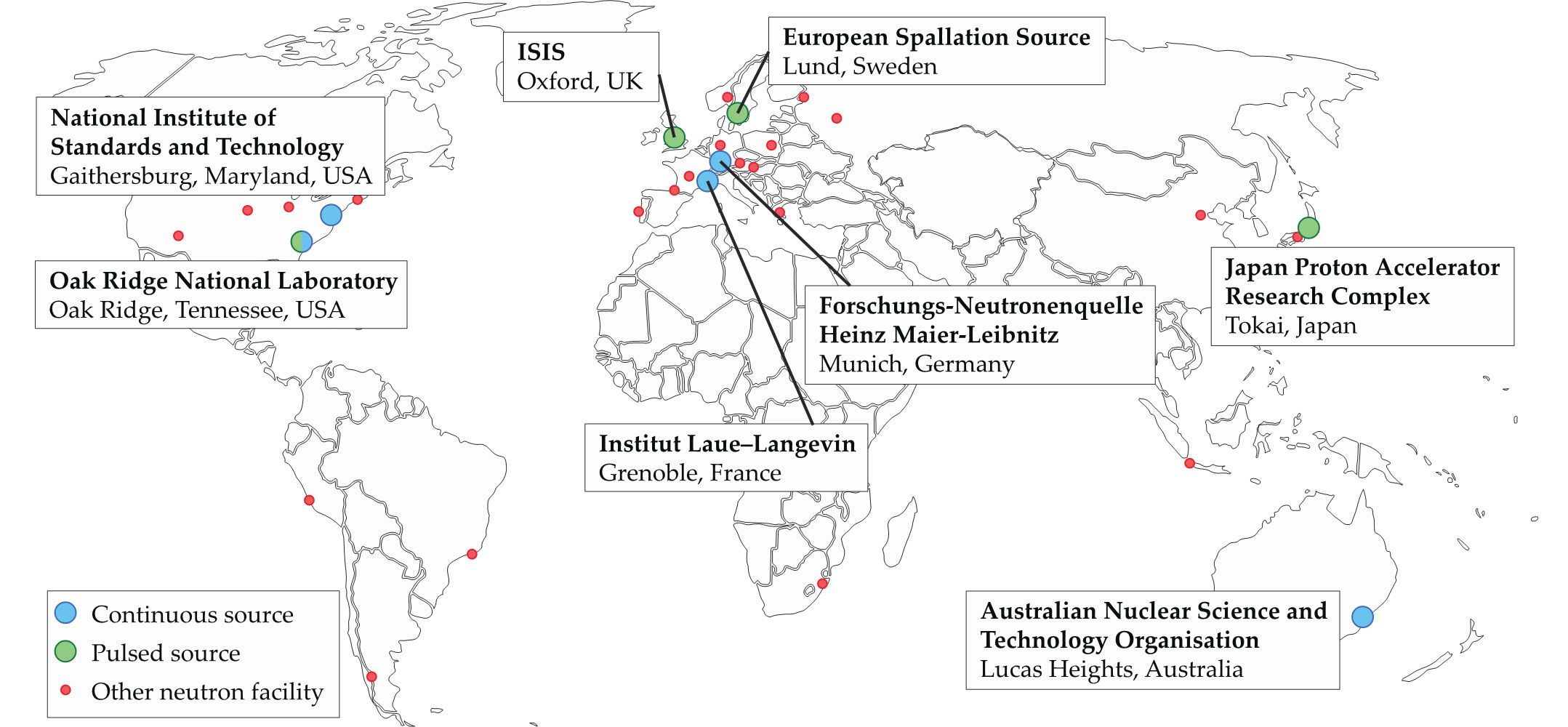
In Europe, the Institut Laue–Langevin (ILL) in Grenoble, France, founded in 1967, pioneered the use of neutrons to study biological systems. Groundbreaking research at the ILL by Heinrich Stuhrmann and Bernard Jacrot drove an increase in the use of neutrons for studying macromolecules in solution. It spurred the establishment of a European Molecular Biology Laboratory (EMBL) outstation at the ILL in 1975 to support the developing field. The Forschungs-Neutronenquelle Heinz Maier-Leibnitz (FRM-II) facility in Germany has also invested considerably in structural biology research. The ISIS Neutron and Muon Source in the UK has focused on the study of soft matter with biological applications. The European Spallation Source (ESS) in Sweden is scheduled to produce its first neutrons in 2020 and should reach full power in 2024; it will be the world’s most powerful spallation source and include several instruments for biological research.
Structural biology initiatives for neutron-based research are also underway in the Asia–Pacific region. The Japan Proton Accelerator Research Complex (J-PARC) spallation source produces high-intensity secondary neutron beams, and the Australian Nuclear Science and Technology Organisation neutron source has a suite of experimental equipment for biological research. The China Spallation Neutron Source, which opened in 2017, is also developing a structural biology program.
Solution scattering
Small-angle neutron scattering (SANS) is the most widely used neutron-based technique for structural biology. It involves diffracting a cold neutron beam with a solution of proteins or other macromolecules, as illustrated in figure
Figure 2.

Small-angle neutron scattering (SANS) probes biomacromolecular structures in solution. (a) In a SANS experiment, the incident beam is scattered at small angles by proteins and other macromolecules in solution. Scattered neutrons are typically collected using a two-dimensional helium ion detector, shown here as a flat screen, and the data are transformed into plots of scattering intensity
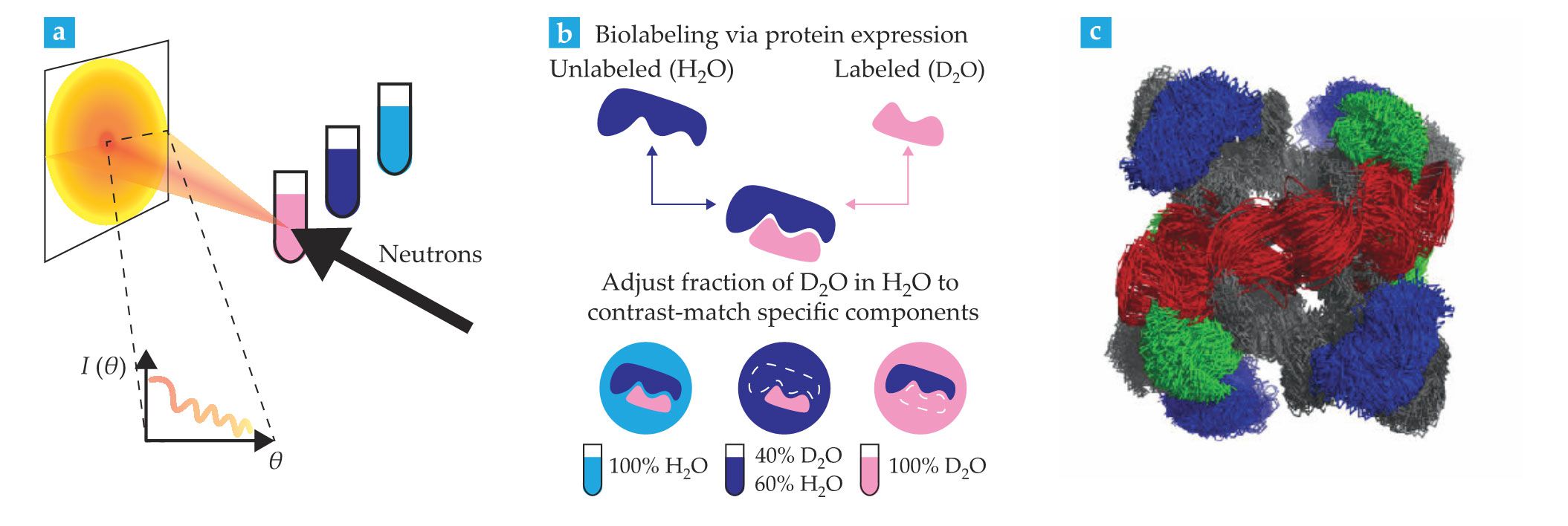
A unique advantage of SANS studies is that they can highlight specific parts of a molecule or molecular complex through solvent contrast variation. The approach, summarized in figure
SANS is sensitive to the relative arrangement of the proteins, nucleic acids, and lipids in assemblies of biological molecules. It can reveal structural arrangements in multicomponent systems and is therefore being increasingly used alongside complementary techniques such as small-angle x-ray scattering (SAXS), NMR, electron microscopy, and mass spectrometry. The three-dimensional structure shown in figure
Stealth nanodisk containers have been developed for use with SANS. They provide a lipid environment into which membrane proteins are inserted. Selective deuteration that renders the containers invisible enables the study of isolated membrane protein structures in a biomimetic lipidic context. 5
More than 20 neutron sources host SANS instruments, and their capabilities for biological research are expanding on multiple fronts. For example, novel instrument and sample configurations have been developed, including integrated size exclusion chromatography and spectroscopy. 6 Anne Martel and collaborators are building an in situ SAXS system on the D11 instrument at the ILL. The combination of SANS and SAXS allows multiple properties of a protein’s structure to be probed simultaneously. A facility having both SANS and SAXS functionalities enables the study of complex biological systems with features on scales ranging from molecular to subcellular.
Emerging computational approaches involving SANS include the development of software programs to reliably evaluate data and to apply advanced molecular dynamics (MD), Monte Carlo, and normal mode analysis methods to building biologically relevant atomistic model structures. 6 , 7 The Small Angle Scattering Biological Data Bank and the BIOISIS repository for SAXS data underpin those approaches by standardizing the deposition and validation of scattering data and models derived from neutron and x-ray sources.
Surface structures
Neutron reflectometry (NR) elucidates the structures of macromolecules on immobilized surfaces. Lipidic systems that mimic biological membranes are of particular interest. As shown in figure
Figure 3.

Neutron reflectometry (NR) is performed on thin films of biological materials prepared on solid, flat substrates. (a) A typical NR sample consists of a lipid bilayer with associated biomolecules such as proteins in a fully hydrated environment, typically a sample container that allows in situ buffer exchange. The fraction of the beam scattered as a specular reflection—with equal incident and reflected angles
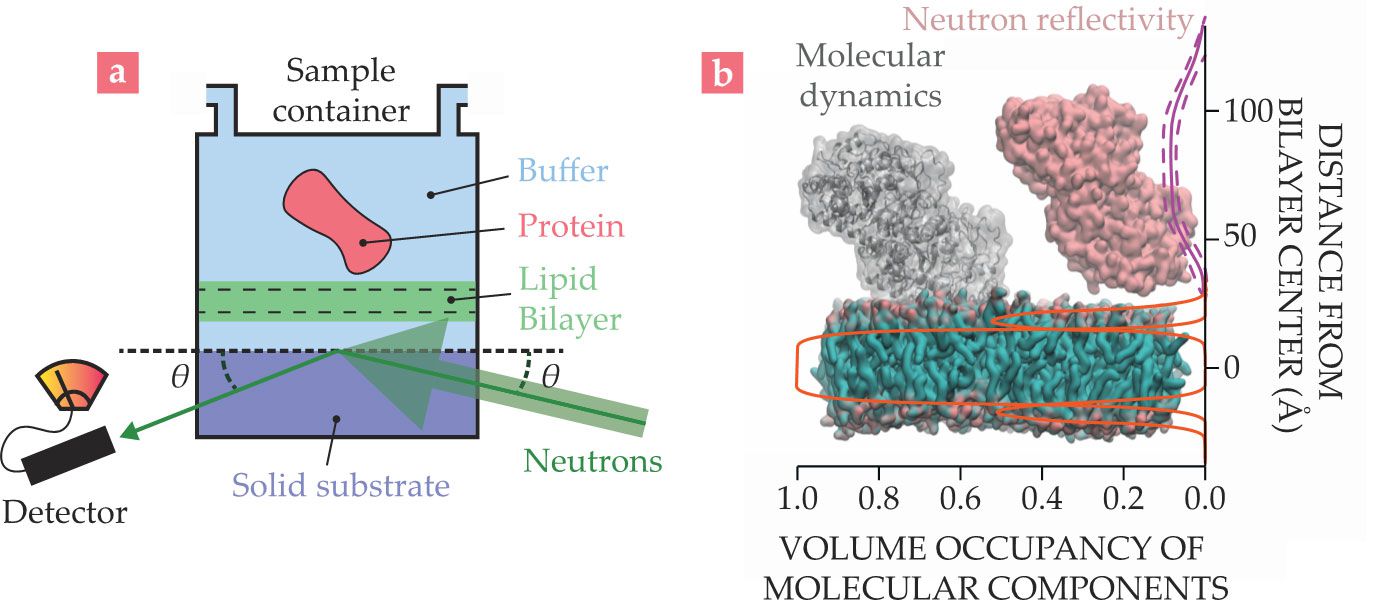
NR is useful for understanding structural arrangements and studying molecular interactions between proteins and membrane macromolecules. Like other neutron methods, it is particularly powerful when combined with other biophysical techniques. For example, NR data collected at NIST in collaboration with the National Institutes of Health were used in combination with MD simulations to understand how the protein tubulin interacts with biological membranes. Tubulin is known for its role as the major component of microtubules in the cytoskeleton, and it is a target for chemotherapeutic drugs such as taxol. However, tubulin has also been shown to associate with outer membranes of mitochondria, which are responsible for cellular respiration.
To understand how that association could occur, researchers analyzed NR data of membrane-bound tubulin using composition space modeling.
9
They described the structure of the membrane–protein complex using information such as molecular volumes, chemical connectivity, and known three-dimensional structures. Figure
At the ILL and ISIS, NR studies have shed light on the relationship between cholesterol and lipid bilayers. By making full use of the ability to produce designer-deuterated analogues of cholesterol and deuterated lipids, the studies have improved our understanding of atherosclerosis. 10
Crystallography
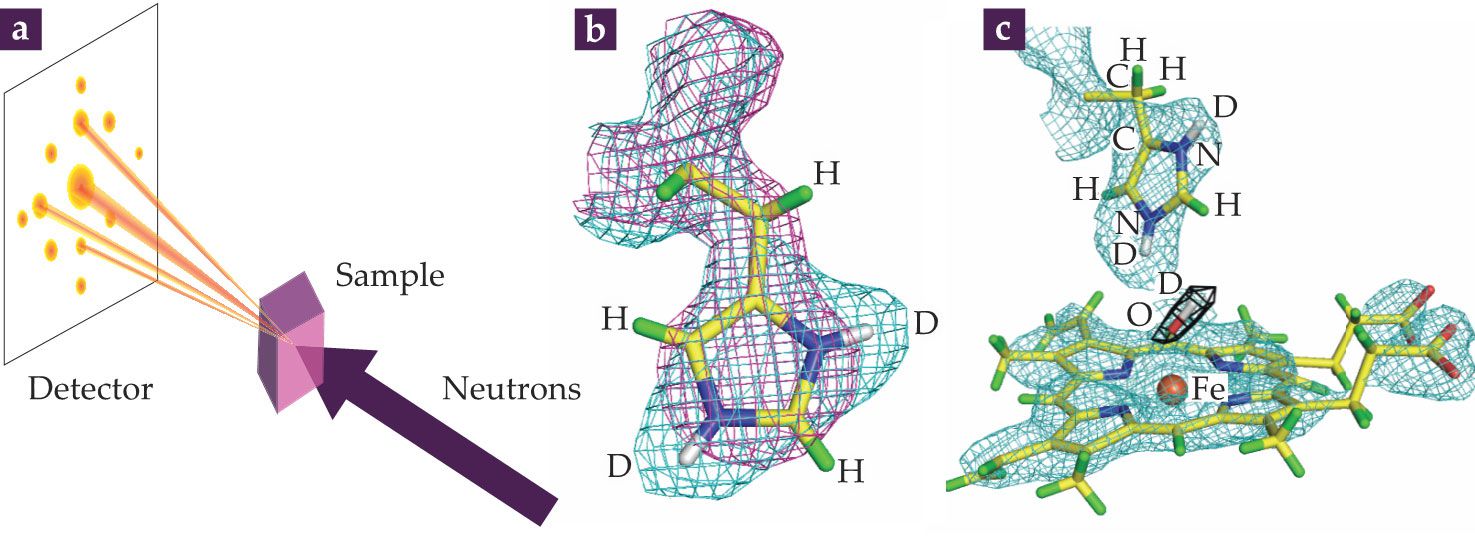
Neutron sources can determine single-crystal protein structures with atomic or near-atomic resolution. The experimental arrangement used for neutron macromolecular crystallography (NMX) is shown in figure
Figure 4.

(a) In neutron macromolecular crystallography (NMX), diffraction data, which are observed as reflections, are collected from a deuterated water–soaked protein crystal sample irradiated by a neutron beam. Reflections are transformed into a nuclear scattering density using phase information, which is often obtained from the x-ray crystal structure of the same protein. (b) The purple mesh shows the electron density of a histidine residue; the cyan mesh shows the corresponding density derived from neutron data. The atomic structure is shown as a stick diagram with hydrogen atoms in green and deuterium in white. The non-overlapping cyan mesh indicates the presence of D atoms that can only be visualized using neutrons. (c) A portion of the crystal structure of ascorbate peroxidase (APX) was determined by combining crystallographic data obtained using x rays and neutrons. The stick diagram shows the atomic structure; cyan mesh shows the density from the neutron map surrounding the iron-containing compound (orange sphere) in APX. The black mesh depicts the nuclear density of an oxygen–deuterium chemical intermediate (red and white stick). Studying the interactions of the O–D intermediate with an Fe-containing compound helps uncover how oxidative enzymes function. (Panel a courtesy of David Hoogerheide; panels b and c adapted by Peter Moody at the University of Leicester, UK, from ref.
The Protein Data Bank archive is an international repository that contains more than 160 000 protein crystal structures. The majority have been determined by x-ray crystallography, which means they do not provide detailed information about the location of H atoms. Only about 150 proteins have been studied using NMX, and most of those studies occurred relatively recently as a result of developments at neutron facilities worldwide.
The lack of NMX-derived information in the data bank is a severe limitation given that H atoms are central to a protein’s structure, function, and interactions. Neutrons directly visualize the locations and orientations of water molecules around biological macromolecules, the protonation of amino acids, and the details of hydrogen bond arrangements (see figure
Peter Moody at the University of Leicester and collaborators have recently done research involving the iron-containing proteins cytochrome c peroxidase (CcP) and ascorbate peroxidase (APX). That work provides an excellent example of how neutron protein crystallography has revealed novel information about the roles of protons and H2O in enzyme catalysis. Those model systems have helped to unravel the mysteries of the degradation of hydrogen peroxide and other small molecules. CcP and APX carry an Fe-containing organic cofactor, known as a heme, buried in their centers. The structural arrangement of the heme is similar to that of hemoglobin, the oxygen-binding protein in blood. To carry out catalytic reactions, the heme in the center of a peroxidase needs to interact with O or an O-containing molecule such as H2O2.
Researchers are intrigued by the nature of Fe–O complexes—both what they look like and where the protons are located in the proteins that help stabilize them. Structures of important Fe–O complexes in CcP have typically been obtained by x-ray crystallography. However, the x rays release electrons that alter the chemical states of metals, including the Fe in heme enzymes. Figure
Improved instrumentation for neutron-based protein crystallography has considerably increased the speed of data acquisition and volume of data that can be obtained using NMX. Cylindrical neutron-sensitive image-plate (NIP) detectors and detector arrays maximize the solid angle of data capture and the efficiency of data acquisition. At the ILL, Matthew Blakeley and colleagues have driven successive upgrades of the LADI-III diffractometer that have delivered improvements in effective neutron flux. It now collects better-quality data with higher throughput. NMX beamlines worldwide use cryocooling to freeze short-lived chemical intermediates in protein crystals. 11 Data from those crystals enable researchers to visualize the atomic structures of unstable protein intermediates, including the locations of H atoms.
Protein dynamics
Protein dynamics such as hinge-bending movements, large-amplitude collective motions, and sampling of different structural arrangements are essential for biological function. 12 Neutron scattering techniques are highly effective for studying motion in biological systems; they are sensitive to length scales ranging from 1 Å to 100 Å and to time scales in the femtosecond to microsecond range.
Experiments typically use solutions of proteins in various solvents that have different levels of deuteration. Protein dynamics can also be studied in living cells. Depending on the experimental configuration, data can be extracted through either incoherent or coherent scattering from samples. Techniques such as quasielastic neutron scattering (QENS) probe H dynamics in proteins by measuring incoherent scattering. Coherent scattering techniques, such as neutron spin echo (NSE) and backscattering spectrometry, detect pair correlations and can be used to study collective dynamics in proteins. 12 For example, NSE has been used to study how two large domains of the enzyme phosphoglycerate kinase move in solution to form a structure that enables it to perform catalysis. 13
Small-angle neutron scattering supplies some information on the assembly of proteins and complexes, and information from neutron spectroscopy can add details about their movements. In the context of other biochemical and biophysical experiments, those data can elucidate macromolecular dynamics and their relationships to biological function in many processes, including enzyme catalysis, protein folding, motions in folded and intrinsically disordered proteins, and fluctuations in the ribosome structure. 13 , 14
Although the number of instruments available for the study of macromolecular dynamics using neutrons is relatively small, all the major facilities have significant capability in the area and are continually upgrading and incorporating emerging technical developments. Upgrades to the ILL’s backscattering and time-of-flight spectrometers have increased their incident flux and energy resolution, thus improving data quality and, crucially, reducing the amount of sample material required. J-PARC has been gradually building its user program and expanding its capability to study macromolecules since 2008; it now has six instruments for inelastic or quasielastic scattering at its Materials and Life Science Experimental Facility.
At FRM-II, inelastic scattering experiments will benefit from planned upgrades, including neutron guides with supermirror coatings and a new time-of-flight spectrometer, increasing the neutron flux to samples. The neutron spin-echo spectrometer at NIST has been upgraded with a new neutron polarization device, which has yielded a significant reduction in data collection time and required sample size while enhancing the range of accessible time scales. In 2023, with collaboration from the Center for Neutron Science at the University of Delaware and funds from NSF, a new spin-echo spectrometer will increase NIST’s data collection rate by an additional order of magnitude.
Emerging capability
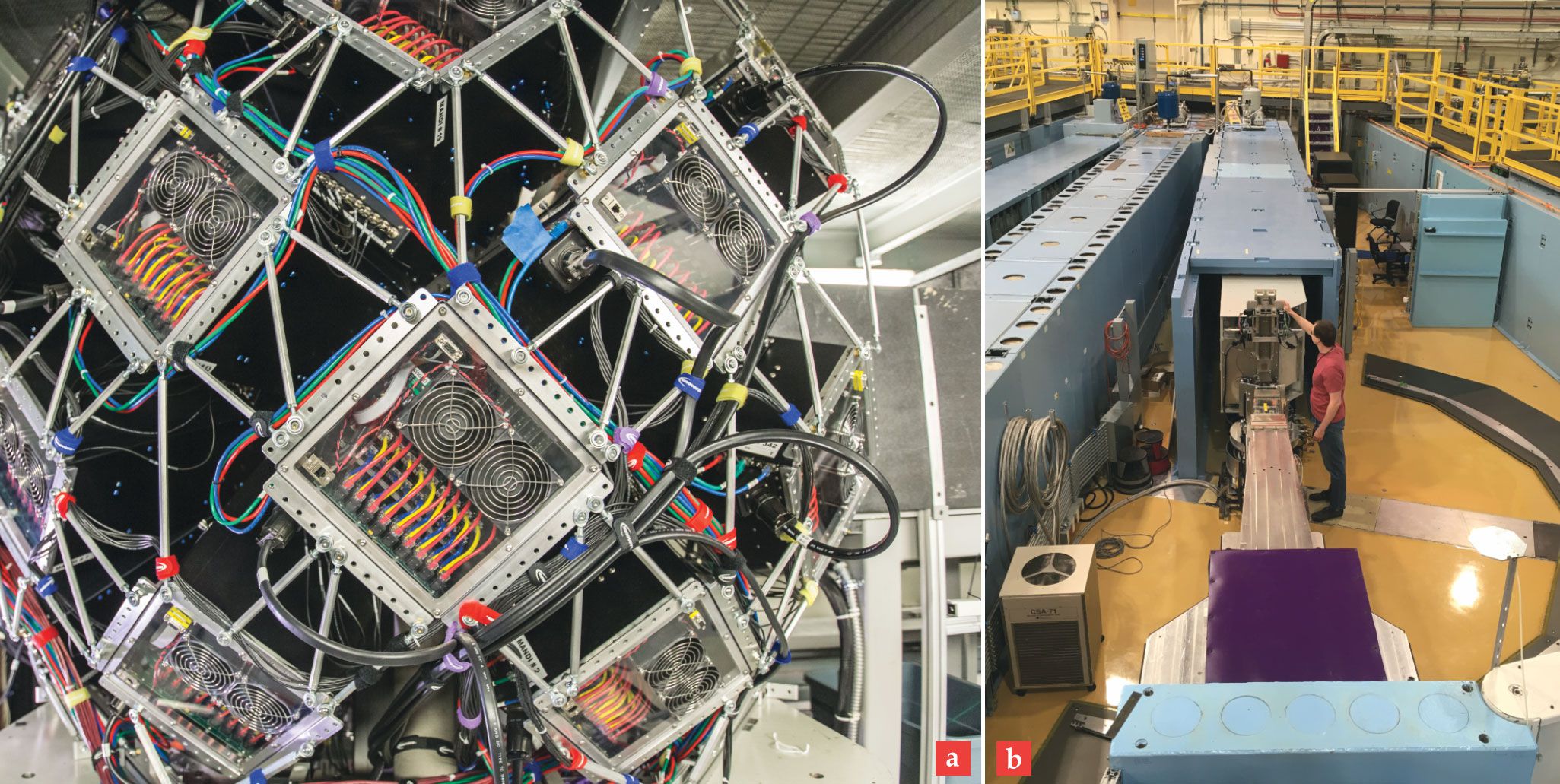
Neutron facilities worldwide are working to increase the range and diversity of possible structure-based studies of biological systems. New diffractometers are being developed to improve NMX and to reduce demand on existing instruments. The ILL is constructing a new instrument called DALI to improve capacity and to allow researchers to study crystals with larger unit cells. New, fast detectors on the Macromolecular Neutron Diffractometer at ORNL (shown in figure
Figure 5.

(a) The large metal framework of the Macromolecular Neutron Diffractometer (MaNDi) at Oak Ridge National Laboratory supports numerous scintillation detectors used to collect diffraction data from protein crystals. (b) The new Chromatic Analysis Neutron Diffractometer or Reflectometer (CANDOR) at the NIST Center for Neutron Research in Gaithersburg, Maryland, discriminates between neutron energies using Bragg diffraction with highly oriented pyrolytic graphite analyzer crystals. MaNDi and CANDOR represent a new generation of instruments for collecting neutron data with increased sensitivity and higher throughput.
(a) GENEVIEVE MARTIN/ORNL (b) DAN NEUMANN/NIST
As throughput increases, so will the need for data portability and standardization across sources and beamlines. The Diffraction Integration for Advanced Light Sources project has developed open-source software to address that need by producing a framework for the collection and analysis of data.
One example of new instrumentation for NR is the Chromatic Analysis Neutron Diffractometer or Reflectometer, which is being commissioned at NIST. Shown in figure
Neutrons have considerable potential for imaging on length scales of multicellular organizations and tissues. They are particularly well-suited to that job because their penetration depth into biological materials can be tuned by changing the irradiation energy. For example, neutron tomography techniques, including emerging methods such as small-angle tensor tomography, 15 offer the possibility of imaging 3D structures in tissues and whole organisms with micrometer resolution. The ESS has identified neutron tomography as a technique that could benefit from its higher brilliance source, which allows for more rapid data collection and improved resolution.
Other recent innovations include axisymmetric mirrors to reflect and focus neutrons. Wolter optics, originally developed for telescopes, have already been implemented in a compact SANS instrument at the Spallation Neutron Source at ORNL. 16 Such setups are opening possibilities for neutron imaging of live biological samples.

A cylindrical detector in the LADI instrument at the Institut Laue–Langevin in Grenoble, France, records neutron diffraction patterns. The data enable the generation of three-dimensional molecular structures that include hydrogen atoms. (Bob Cubitt/ILL.)

Strategies and collaboration
In July 2018 the American Physical Society’s Panel on Public Affairs published a report, Neutrons for the Nation. The 32-page report highlights current and future issues around the need to maintain national facilities supporting R&D that involves neutrons. Such activities are “essential components of R&D in numerous areas of science and engineering,” the report says. “Neutron sources play a key role in overall U.S. innovation capacity.”
The report also points to a decline in neutron R&D efforts in the US. It acknowledges an urgent need to invest in maintaining diverse and complementary capabilities, including spallation facilities, research reactors, and high-performance instrumentation, as part of reestablishing the US as a world leader in neutron research. In Europe, the League of Advanced European Neutron Sources was created to coordinate efforts to increase the impact of neutron science. The key aim of the league’s charter, signed in September 2018, is to “facilitate any form of discussion and decision-making process that has the potential to strengthen European neutron science via enhanced collaboration among the facilities.”
Well-defined strategies and cooperation between facilities at national and international levels will help maximize the potential for using neutrons to study biological problems. An emerging trend among large-scale facilities is to give users access to multiple techniques, such as state-of-the-art neutron, x-ray, cryoelectron microscopy, and NMR platforms, at or near the same site, as now routinely happens at the Grenoble Partnership for Structural Biology. Biological neutron scattering is at its most powerful when combined with other techniques that provide unique insights into the structure, assembly, and function of macromolecules. Such collaboration presents a great opportunity for physicists to engage with challenging problems in biological research to impact both fundamental knowledge and human health.
References
1. V. T. Forsyth, P. Moody, Acta Crystallogr., Sect. D: Biol. Crystallogr. 74, 1126 (2018); https://doi.org/10.1107/S2059798318017886
R. Ashkar et al., Acta Crystallogr., Sect. D: Biol. Crystallogr. 74, 1129 (2018); https://doi.org/10.1107/S2059798318017503
G. Zaccai, Eur. Biophys. J. 41, 781 (2012). https://doi.org/10.1007/s00249-012-0825-52. J. H. Lakey, J. R. Soc. Interface 6, S567 (2009); https://doi.org/10.1098/rsif.2009.0156.focus
R. Schneider et al., J. Mol. Biol. 41, 231 (1969); https://doi.org/10.1016/0022-2836(69)90388-X
H. B. Stuhrmann, J. Appl. Crystallogr. 7, 173 (1974); https://doi.org/10.1107/S0021889874009071
B. P. Schoenborn, Nature 224, 143 (1969); https://doi.org/10.1038/224143a0
B. Jacrot, Rep. Prog. Phys. 39, 911 (1976). https://doi.org/10.1088/0034-4885/39/10/0013. M. Haertlein et al., in Isotope Labeling of Biomolecules—Applications, Z. Kelman, ed., Academic Press/Elsevier (2016), p. 113.
4. A. Lapinaite et al., Nature 502, 519 (2013). https://doi.org/10.1038/nature12581
5. S. Maric et al., Acta Crystallogr., Sect. D: Biol. Crystallogr. 70, 317 (2014); https://doi.org/10.1107/S1399004713027466
I. Josts et al., Structure 26, 1072 (2018); https://doi.org/10.1016/j.str.2018.05.007
J. Nitsche et al., Commun. Biol. 1, 206 (2018). https://doi.org/10.1038/s42003-018-0203-76. E. Mahieu, F. Gabel, Acta Crystallogr., Sect. D: Biol. Crystallogr. 74, 715 (2018); https://doi.org/10.1107/S2059798318005016
J. Trewhella, in Integrative Structural Biology with Hybrid Methods, H. Nakamura et al., eds., Springer (2018), p. 77;
A. Jordan et al., J. Appl. Crystallogr. 49, 2015 (2016); https://doi.org/10.1107/S1600576716016514
E. Valentini et al., Nucleic Acids Res. 43, D357 (2015). https://doi.org/10.1093/nar/gku10477. S. Grudinin, M. Garkavenko, A. Kazennov, Acta Crystallogr., Sect. D: Biol. Crystallogr. 73, 449 (2017). https://doi.org/10.1107/S2059798317005745
8. C. F. Majkrzak et al., in Neutron Scattering in Biology: Techniques and Applications, J. Fitter, T. Gutberlet, J. Katsaras, eds., Springer (2006), p. 225.
9. P. Shekhar et al., J. Appl. Phys. 110, 102216 (2011); https://doi.org/10.1063/1.3661986
D. P. Hoogerheide et al., Proc. Natl. Acad. Sci. USA 114, E3622 (2017). https://doi.org/10.1073/pnas.161980611410. S. Waldie et al., Langmuir 34, 472 (2018); https://doi.org/10.1021/acs.langmuir.7b02716
M. Moulin et al., Chem. Phys. Lipids 212, 80 (2018); https://doi.org/10.1016/j.chemphyslip.2018.01.006
A. Luchini et al., Colloids Surf. B 168, 126 (2018); https://doi.org/10.1016/j.colsurfb.2018.02.009
S. Waldie et al., Sci. Rep. 9, 5118 (2019). https://doi.org/10.1038/s41598-019-41439-z11. C. M. Casadei et al., Science 345, 193 (2014); https://doi.org/10.1126/science.1254398
H. Kwon et al., Nat. Commun. 7, 13445 (2016); https://doi.org/10.1038/ncomms13445
M. G. Cuypers et al., Sci. Rep. 6, 31487 (2016); https://doi.org/10.1038/srep31487
A. W. Yee et al., Nat. Commun. 10, 925 (2019); https://doi.org/10.1038/s41467-019-08609-z
G. Zaccai, Biochim. Biophys. Acta 1864, 129475 (2020). https://doi.org/10.1016/j.bbagen.2019.12947512. J. C. Smith et al., Annu. Rev. Biophys. 47, 335 (2018). https://doi.org/10.1146/annurev-biophys-070317-033358
13. R. Inoue et al., Biophys. J. 99, 2309 (2010). https://doi.org/10.1016/j.bpj.2010.08.017
14. M. R. Duff Jr et al., Biochemistry 57, 4263 (2018); https://doi.org/10.1021/acs.biochem.8b00424
F. Ameseder et al., Phys. Chem. Chem. Phys. 20, 5128 (2018); https://doi.org/10.1039/C7CP08292D
G. Schirò et al., Nat. Commun. 6, 6490 (2015); https://doi.org/10.1038/ncomms7490
G. Zaccai et al., Sci. Rep. 6, 37138 (2016). https://doi.org/10.1038/srep3713815. M. Liebi et al., Nature 527, 349 (2015). https://doi.org/10.1038/nature16056
16. D. Liu et al., Nat. Commun. 4, 2556 (2013). https://doi.org/10.1038/ncomms3556
More about the authors
David Hoogerheide is a researcher at the National Institute of Standards and Technology Center for Neutron Research in Gaithersburg, Maryland. Trevor Forsyth is a senior fellow and head of the life sciences group at the Institut Laue–Langevin in Grenoble, France; he also holds a chair in biophysics at Keele University in the UK. Katherine Brown is a senior researcher in the Cavendish Laboratory at Cambridge University in the UK and at the University of Texas at Austin.







